SF3B1 mutations in chronic lymphocytic leukemia
- PMID: 23568491
- PMCID: PMC3674664
- DOI: 10.1182/blood-2013-02-427641
SF3B1 mutations in chronic lymphocytic leukemia
Abstract
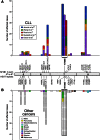
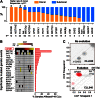
SF3B1 is a critical component of the splicing machinery, which catalyzes the removal of introns from precursor messenger RNA (mRNA). Next-generation sequencing studies have identified mutations in SF3B1 in chronic lymphocytic leukemia (CLL) at high frequency. In CLL, SF3B1 mutation is associated with more aggressive disease and shorter survival, and recent studies suggest that it can be incorporated into prognostic schema to improve the prediction of disease progression. Mutations in SF3B1 are predominantly subclonal genetic events in CLL, and hence are likely later events in the progression of CLL. Evidence of altered pre-mRNA splicing has been detected in CLL cases with SF3B1 mutations. Although the causative link between SF3B1 mutation and CLL pathogenesis remains unclear, several lines of evidence suggest SF3B1 mutation might be linked to genomic stability and epigenetic modification.
Figures


References
-
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. - PubMed
-
- Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44(1):47–52. - PubMed
-
- Mansouri L, Cahill N, Gunnarsson R, et al. NOTCH1 and SF3B1 mutations can be added to the hierarchical prognostic classification in chronic lymphocytic leukemia. Leukemia. 2013;27(2):512–514. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

