Country-specific antibiotic use practices impact the human gut resistome
- PMID: 23568836
- PMCID: PMC3698509
- DOI: 10.1101/gr.155465.113
Country-specific antibiotic use practices impact the human gut resistome
Abstract
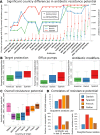
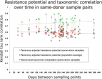
Despite increasing concerns over inappropriate use of antibiotics in medicine and food production, population-level resistance transfer into the human gut microbiota has not been demonstrated beyond individual case studies. To determine the "antibiotic resistance potential" for entire microbial communities, we employ metagenomic data and quantify the totality of known resistance genes in each community (its resistome) for 68 classes and subclasses of antibiotics. In 252 fecal metagenomes from three countries, we show that the most abundant resistance determinants are those for antibiotics also used in animals and for antibiotics that have been available longer. Resistance genes are also more abundant in samples from Spain, Italy, and France than from Denmark, the United States, or Japan. Where comparable country-level data on antibiotic use in both humans and animals are available, differences in these statistics match the observed resistance potential differences. The results are robust over time as the antibiotic resistance determinants of individuals persist in the human gut flora for at least a year.
Figures

References
-
- Aarestrup F 2012. Sustainable farming: Get pigs off antibiotics. Nature 486: 465–466 - PubMed
-
- Agersø Y, Lester CH, Porsbo LJ, Orsted I, Emborg H-D, Olsen KEP, Jensen LB, Heuer OE, Frimodt-Møller N, Aarestrup FM, et al. 2008. Vancomycin-resistant Enterococcus faecalis isolates from a Danish patient and two healthy human volunteers are possibly related to isolates from imported turkey meat. J Antimicrob Chemother 62: 844–845 - PubMed
-
- Bager F, Madsen M, Christensen J, Aarestrup FM 1997. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev Vet Med 31: 95–112 - PubMed
-
- Barbosa TM, Levy SB 2000. The impact of antibiotic use on resistance development and persistence. Drug Resist Updat 3: 303–311 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
