Cancer translocations in human cells induced by zinc finger and TALE nucleases
- PMID: 23568838
- PMCID: PMC3698511
- DOI: 10.1101/gr.147314.112
Cancer translocations in human cells induced by zinc finger and TALE nucleases
Abstract
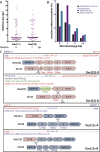
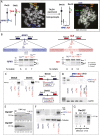
Chromosomal translocations are signatures of numerous cancers and lead to expression of fusion genes that act as oncogenes. The wealth of genomic aberrations found in cancer, however, makes it challenging to assign a specific phenotypic change to a specific aberration. In this study, we set out to use genome editing with zinc finger (ZFN) and transcription activator-like effector (TALEN) nucleases to engineer, de novo, translocation-associated oncogenes at cognate endogenous loci in human cells. Using ZFNs and TALENs designed to cut precisely at relevant translocation breakpoints, we induced cancer-relevant t(11;22)(q24;q12) and t(2;5)(p23;q35) translocations found in Ewing sarcoma and anaplastic large cell lymphoma (ALCL), respectively. We recovered both translocations with high efficiency, resulting in the expression of the EWSR1-FLI1 and NPM1-ALK fusions. Breakpoint junctions recovered after ZFN cleavage in human embryonic stem (ES) cell-derived mesenchymal precursor cells fully recapitulated the genomic characteristics found in tumor cells from Ewing sarcoma patients. This approach with tailored nucleases demonstrates that expression of fusion genes found in cancer cells can be induced from the native promoter, allowing interrogation of both the underlying mechanisms and oncogenic consequences of tumor-related translocations in human cells. With an analogous strategy, the ALCL translocation was reverted in a patient cell line to restore the integrity of the two participating chromosomes, further expanding the repertoire of genomic rearrangements that can be engineered by tailored nucleases.
Figures


References
-
- Chansky HA, Barahmand-Pour F, Mei Q, Kahn-Farooqi W, Zielinska-Kwiatkowska A, Blackburn M, Chansky K, Conrad EU III, Bruckner JD, Greenlee TK, et al. 2004. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing’s sarcoma cells in vitro. J Orthop Res 22: 910–917 - PubMed
-
- Colombo E, Martinelli P, Zamponi R, Shing DC, Bonetti P, Luzi L, Volorio S, Bernard L, Pruneri G, Alcalay M, et al. 2006. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res 66: 3044–3050 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
