An active ac/ds transposon system for activation tagging in tomato cultivar m82 using clonal propagation
- PMID: 23569107
- PMCID: PMC3641199
- DOI: 10.1104/pp.113.213876
An active ac/ds transposon system for activation tagging in tomato cultivar m82 using clonal propagation
Abstract
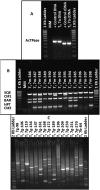
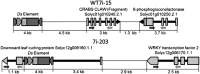
Tomato (Solanum lycopersicum) is a model organism for Solanaceae in both molecular and agronomic research. This project utilized Agrobacterium tumefaciens transformation and the transposon-tagging construct Activator (Ac)/Dissociator (Ds)-ATag-Bar_gosGFP to produce activation-tagged and knockout mutants in the processing tomato cultivar M82. The construct carried hygromycin resistance (hyg), green fluorescent protein (GFP), and the transposase (TPase) of maize (Zea mays) Activator major transcript X054214.1 on the stable Ac element, along with a 35S enhancer tetramer and glufosinate herbicide resistance (BAR) on the mobile Ds-ATag element. An in vitro propagation strategy was used to produce a population of 25 T0 plants from a single transformed plant regenerated in tissue culture. A T1 population of 11,000 selfed and cv M82 backcrossed progeny was produced from the functional T0 line. This population was screened using glufosinate herbicide, hygromycin leaf painting, and multiplex polymerase chain reaction (PCR). Insertion sites of transposed Ds-ATag elements were identified through thermal asymmetric interlaced PCR, and resulting product sequences were aligned to the recently published tomato genome. A population of 509 independent, Ds-only transposant lines spanning all 12 tomato chromosomes has been developed. Insertion site analysis demonstrated that more than 80% of these lines harbored Ds insertions conducive to activation tagging. The capacity of the Ds-ATag element to alter transcription was verified by quantitative real-time reverse transcription-PCR in two mutant lines. The transposon-tagged lines have been immortalized in seed stocks and can be accessed through an online database, providing a unique resource for tomato breeding and analysis of gene function in the background of a commercial tomato cultivar.
Figures



Similar articles
-
Identification and isolation of the FEEBLY gene from tomato by transposon tagging.Mol Gen Genet. 1996 Jun 12;251(3):267-80. doi: 10.1007/BF02172517. Mol Gen Genet. 1996. PMID: 8676869
-
Analysis of the frequency of inheritance of transposed Ds elements in Arabidopsis after activation by a CaMV 35S promoter fusion to the Ac transposase gene.Mol Gen Genet. 1993 Dec;241(5-6):627-36. doi: 10.1007/BF00279905. Mol Gen Genet. 1993. PMID: 8264537
-
An Efficient System for Ds Transposon Tagging in Brachypodium distachyon.Plant Physiol. 2019 May;180(1):56-65. doi: 10.1104/pp.18.00875. Epub 2019 Mar 13. Plant Physiol. 2019. PMID: 30867334 Free PMC article.
-
DNA methylation and activity of the maize Spm transposable element.Curr Top Microbiol Immunol. 1995;197:143-64. doi: 10.1007/978-3-642-79145-1_10. Curr Top Microbiol Immunol. 1995. PMID: 7493489 Review. No abstract available.
-
Molecular biology of maize Ac/Ds elements: an overview.Methods Mol Biol. 2013;1057:59-82. doi: 10.1007/978-1-62703-568-2_5. Methods Mol Biol. 2013. PMID: 23918421 Review.
Cited by
-
A Tomato EMS-Mutagenized Population Provides New Valuable Resources for Gene Discovery and Breeding of Developmental Traits.Plants (Basel). 2022 Sep 20;11(19):2453. doi: 10.3390/plants11192453. Plants (Basel). 2022. PMID: 36235319 Free PMC article.
-
High-throughput generation of an activation-tagged mutant library for functional genomic analyses in tobacco.Planta. 2015 Mar;241(3):629-40. doi: 10.1007/s00425-014-2186-z. Epub 2014 Nov 19. Planta. 2015. PMID: 25408504
-
Development of activation-tagged gain-of-functional mutants in indica rice line (BPT 5204) for sheath blight resistance.Mol Biol Rep. 2024 Mar 2;51(1):381. doi: 10.1007/s11033-023-09194-7. Mol Biol Rep. 2024. PMID: 38430361
-
Different Fruit-Specific Promoters Drive AtMYB12 Expression to Improve Phenylpropanoid Accumulation in Tomato.Molecules. 2022 Jan 5;27(1):317. doi: 10.3390/molecules27010317. Molecules. 2022. PMID: 35011551 Free PMC article.
-
Transposon based activation tagging in diploid strawberry and monoploid derivatives of potato.Plant Cell Rep. 2014 Jul;33(7):1203-16. doi: 10.1007/s00299-014-1610-y. Epub 2014 Apr 12. Plant Cell Rep. 2014. PMID: 24728112
References
-
- Aboul-Soud MAM, Chen X, Kang JG, Yun BW, Raja MU, Malik SI, Loake GJ. (2009) Activation tagging of ADR2 conveys a spreading lesion phenotype and resistance to biotrophic pathogens. New Phytol 183: 1163–1175 - PubMed
-
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. (2011) The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res 39: D1149–D1155 - PMC - PubMed
-
- Bríza J, Niedermeierová H, Pavingerová D, Thomas CM, Klimyuk VI, Jones JDG. (2002) Transposition patterns of unlinked transposed Ds elements from two T-DNA loci on tomato chromosomes 7 and 8. Mol Genet Genomics 266: 882–890 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

