Differential protein partitioning within the herpesvirus tegument and envelope underlies a complex and variable virion architecture
- PMID: 23569236
- PMCID: PMC3637715
- DOI: 10.1073/pnas.1221896110
Differential protein partitioning within the herpesvirus tegument and envelope underlies a complex and variable virion architecture
Abstract
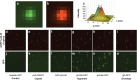
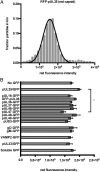
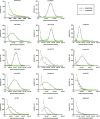
The herpesvirus virion is a multilayered structure consisting of a DNA-filled capsid, tegument, and envelope. Detailed reconstructions of the capsid are possible based on its icosahedral symmetry, but the surrounding tegument and envelope layers lack regular architecture. To circumvent limitations of symmetry-based ultrastructural reconstruction methods, a fluorescence approach was developed using single-particle imaging combined with displacement measurements at nanoscale resolution. An analysis of 11 tegument and envelope proteins defined the composition and plasticity of symmetric and asymmetric elements of the virion architecture. The resulting virion protein map ascribes molecular composition to density profiles previously acquired by traditional ultrastructural methods, and provides a way forward to examine the dynamics of the virion architecture during infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

