BioCode: two biologically compatible Algorithms for embedding data in non-coding and coding regions of DNA
- PMID: 23570444
- PMCID: PMC3698116
- DOI: 10.1186/1471-2105-14-121
BioCode: two biologically compatible Algorithms for embedding data in non-coding and coding regions of DNA
Abstract
Background: In recent times, the application of deoxyribonucleic acid (DNA) has diversified with the emergence of fields such as DNA computing and DNA data embedding. DNA data embedding, also known as DNA watermarking or DNA steganography, aims to develop robust algorithms for encoding non-genetic information in DNA. Inherently DNA is a digital medium whereby the nucleotide bases act as digital symbols, a fact which underpins all bioinformatics techniques, and which also makes trivial information encoding using DNA straightforward. However, the situation is more complex in methods which aim at embedding information in the genomes of living organisms. DNA is susceptible to mutations, which act as a noisy channel from the point of view of information encoded using DNA. This means that the DNA data embedding field is closely related to digital communications. Moreover it is a particularly unique digital communications area, because important biological constraints must be observed by all methods. Many DNA data embedding algorithms have been presented to date, all of which operate in one of two regions: non-coding DNA (ncDNA) or protein-coding DNA (pcDNA).
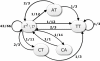
Results: This paper proposes two novel DNA data embedding algorithms jointly called BioCode, which operate in ncDNA and pcDNA, respectively, and which comply fully with stricter biological restrictions. Existing methods comply with some elementary biological constraints, such as preserving protein translation in pcDNA. However there exist further biological restrictions which no DNA data embedding methods to date account for. Observing these constraints is key to increasing the biocompatibility and in turn, the robustness of information encoded in DNA.
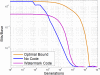
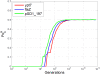
Conclusion: The algorithms encode information in near optimal ways from a coding point of view, as we demonstrate by means of theoretical and empirical (in silico) analyses. Also, they are shown to encode information in a robust way, such that mutations have isolated effects. Furthermore, the preservation of codon statistics, while achieving a near-optimum embedding rate, implies that BioCode pcDNA is also a near-optimum first-order steganographic method.
Figures



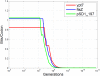


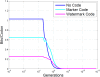
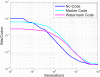

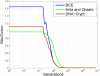
References
-
- Wong PC, Wong K, Foote H. Organic data memory using the DNA, approach. Comms ACM. 2003;46:95–98.
-
- Shimanovsky B, Feng J, Potkonjak M. Procs. of the 5th Intl. Workshop in Information Hiding Noordwijkerhout. The Netherlands; 2002. Hiding data in DNA; pp. 373–386.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

