Low penetrance susceptibility to glioma is caused by the TP53 variant rs78378222
- PMID: 23571737
- PMCID: PMC3670481
- DOI: 10.1038/bjc.2013.155
Low penetrance susceptibility to glioma is caused by the TP53 variant rs78378222
Abstract
Background: Most of the heritable risk of glioma is presently unaccounted for by mutations in known genes. In addition to rare inactivating germline mutations in TP53 causing glioma in the context of the Li-Fraumeni syndrome, polymorphic variation in TP53 may also contribute to the risk of developing glioma.
Methods: To comprehensively evaluate the impact of variation in TP53 on risk, we analysed 23 tagSNPs and imputed 2377 unobserved genotypes in four series totaling 4147 glioma cases and 7435 controls.
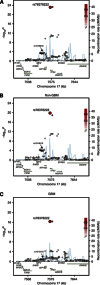
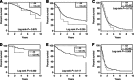
Results: The strongest validated association signal was shown by the imputed single-nucleotide polymorphism (SNP) rs78378222 (P=6.86 × 10(-24), minor allele frequency ~0.013). Confirmatory genotyping confirmed the high quality of the imputation. The association between rs78378222 and risk was seen for both glioblastoma multiforme (GBM) and non-GBM tumours. We comprehensively examined the relationship between rs78378222 and overall survival in two of the case series totaling 1699 individuals. Despite employing statistical tests sensitive to the detection of differences in early survival, no association was shown.
Conclusion: Our data provided strong validation of rs78378222 as a risk factor for glioma but do not support the tenet that the polymorphism being a clinically useful prognostic marker. Acquired TP53 inactivation is a common feature of glioma. As rs78378222 changes the polyadenylation signal of TP53 leading to impaired 3'-end processing of TP53 mRNA, the SNP has strong plausibility for being directly functional contributing to the aetiological basis of glioma.
Figures


References
-
- Bondy ML, Scheurer ME, Malmer B, Barnholtz-Sloan JS, Davis FG, Il'yasova D, Kruchko C, McCarthy BJ, Rajaraman P, Schwartzbaum JA, Sadetzki S, Schlehofer B, Tihan T, Wiemels JL, Wrensch M, Buffler PA. Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008;113:1953–1968. - PMC - PubMed
-
- Cardis E, Richardson L, Deltour I, Armstrong B, Feychting M, Johansen C, Kilkenny M, McKinney P, Modan B, Sadetzki S, Schuz J, Swerdlow A, Vrijheid M, Auvinen A, Berg G, Blettner M, Bowman J, Brown J, Chetrit A, Christensen HC, Cook A, Hepworth S, Giles G, Hours M, Iavarone I, Jarus-Hakak A, Klaeboe L, Krewski D, Lagorio S, Lonn S, Mann S, McBride M, Muir K, Nadon L, Parent ME, Pearce N, Salminen T, Schoemaker M, Schlehofer B, Siemiatycki J, Taki M, Takebayashi T, Tynes T, van Tongeren M, Vecchia P, Wiart J, Woodward A, Yamaguchi N. The INTERPHONE study: design, epidemiological methods, and description of the study population. Eur J Epidemiol. 2007;22:647–664. - PubMed
-
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. - PubMed
-
- Hemminki K, Tretli S, Sundquist J, Johannesen TB, Granstrom C. Familial risks in nervous-system tumours: a histology-specific analysis from Sweden and Norway. Lancet Oncol. 2009;10:481–488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

