Genomic prediction in CIMMYT maize and wheat breeding programs
- PMID: 23572121
- PMCID: PMC3860161
- DOI: 10.1038/hdy.2013.16
Genomic prediction in CIMMYT maize and wheat breeding programs
Abstract
Genomic selection (GS) has been implemented in animal and plant species, and is regarded as a useful tool for accelerating genetic gains. Varying levels of genomic prediction accuracy have been obtained in plants, depending on the prediction problem assessed and on several other factors, such as trait heritability, the relationship between the individuals to be predicted and those used to train the models for prediction, number of markers, sample size and genotype × environment interaction (GE). The main objective of this article is to describe the results of genomic prediction in International Maize and Wheat Improvement Center's (CIMMYT's) maize and wheat breeding programs, from the initial assessment of the predictive ability of different models using pedigree and marker information to the present, when methods for implementing GS in practical global maize and wheat breeding programs are being studied and investigated. Results show that pedigree (population structure) accounts for a sizeable proportion of the prediction accuracy when a global population is the prediction problem to be assessed. However, when the prediction uses unrelated populations to train the prediction equations, prediction accuracy becomes negligible. When genomic prediction includes modeling GE, an increase in prediction accuracy can be achieved by borrowing information from correlated environments. Several questions on how to incorporate GS into CIMMYT's maize and wheat programs remain unanswered and subject to further investigation, for example, prediction within and between related bi-parental crosses. Further research on the quantification of breeding value components for GS in plant breeding populations is required.
Figures
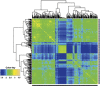
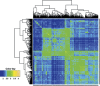

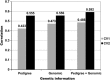
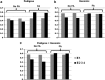
References
-
- Bernardo R. Molecular markers and selection of complex traits in plants: learning from the last 20 years. Crop Sci. 2008;48:1649–1664.
-
- Bernardo R, Yu Y. Prospects for genome-wide selection for quantitative traits in maize. Crop Sci. 2007;47:1082–1090.
-
- Burgueño J, Crossa J, Cornelius PL, Trethowan R, McLaren G, Krishnamachari A. Modeling additive × environment and additive × additive × environment using genetic covariances of relatives of wheat genotypes. Crop Sci. 2007;43:311–320.
-
- Burgueño J, Crossa J, Cotes JM, San Vicente F, Das B. Prediction assessment of linear mixed models for multienvironment trials. Crop Sci. 2011;51:944–954.
-
- Burgueño J, de los Campos GDL, Weigel K, Crossa J. Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Sci. 2012;52:707–719.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

