Identification of rodent homologs of hepatitis C virus and pegiviruses
- PMID: 23572554
- PMCID: PMC3622934
- DOI: 10.1128/mBio.00216-13
Identification of rodent homologs of hepatitis C virus and pegiviruses
Abstract
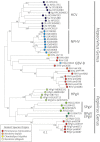
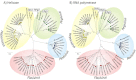
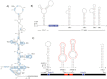
Hepatitis C virus (HCV) and human pegivirus (HPgV or GB virus C) are globally distributed and infect 2 to 5% of the human population. The lack of tractable-animal models for these viruses, in particular for HCV, has hampered the study of infection, transmission, virulence, immunity, and pathogenesis. To address this challenge, we searched for homologous viruses in small mammals, including wild rodents. Here we report the discovery of several new hepaciviruses (HCV-like viruses) and pegiviruses (GB virus-like viruses) that infect wild rodents. Complete genome sequences were acquired for a rodent hepacivirus (RHV) found in Peromyscus maniculatus and a rodent pegivirus (RPgV) found in Neotoma albigula. Unique genomic features and phylogenetic analyses confirmed that these RHV and RPgV variants represent several novel virus species in the Hepacivirus and Pegivirus genera within the family Flaviviridae. The genetic diversity of the rodent hepaciviruses exceeded that observed for hepaciviruses infecting either humans or non-primates, leading to new insights into the origin, evolution, and host range of hepaciviruses. The presence of genes, encoded proteins, and translation elements homologous to those found in human hepaciviruses and pegiviruses suggests the potential for the development of new animal systems with which to model HCV pathogenesis, vaccine design, and treatment.
Importance: The genetic and biological characterization of animal homologs of human viruses provides insights into the origins of human infections and enhances our ability to study their pathogenesis and explore preventive and therapeutic interventions. Horses are the only reported host of nonprimate homologs of hepatitis C virus (HCV). Here, we report the discovery of HCV-like viruses in wild rodents. The majority of HCV-like viruses were found in deer mice (Peromyscus maniculatus), a small rodent used in laboratories to study viruses, including hantaviruses. We also identified pegiviruses in rodents that are distinct from the pegiviruses found in primates, bats, and horses. These novel viruses may enable the development of small-animal models for HCV, the most common infectious cause of liver failure and hepatocellular carcinoma after hepatitis B virus, and help to explore the health relevance of the highly prevalent human pegiviruses.
Figures

References
-
- Berg T, Müller AR, Platz KP, Höhne M, Bechstein WO, Hopf U, Wiedenmann B, Neuhaus P, Schreier E. 1999. Dynamics of GB virus C viremia early after orthotopic liver transplantation indicates extrahepatic tissues as the predominant site of GB virus C replication. Hepatology 29:245–249 - PubMed
-
- Blair CS, Davidson F, Lycett C, McDonald DM, Haydon GH, Yap PL, Hayes PC, Simmonds P, Gillon J. 1998. Prevalence, incidence, and clinical characteristics of hepatitis G virus/GB virus C infection in Scottish blood donors. J. Infect. Dis. 178:1779–1782 - PubMed
-
- Williams CF, Klinzman D, Yamashita TE, Xiang J, Polgreen PM, Rinaldo C, Liu C, Phair J, Margolick JB, Zdunek D, Hess G, Stapleton JT. 2004. Persistent GB virus C infection and survival in HIV-infected men. N. Engl. J. Med. 350:981–990 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R01 AI072613/AI/NIAID NIH HHS/United States
- R21 AI081132/AI/NIAID NIH HHS/United States
- AI090055/AI/NIAID NIH HHS/United States
- AI070411/AI/NIAID NIH HHS/United States
- U01 AI070411/AI/NIAID NIH HHS/United States
- 095831/Wellcome Trust/United Kingdom
- R01 AI090055/AI/NIAID NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- AI081132/AI/NIAID NIH HHS/United States
- AI072613/AI/NIAID NIH HHS/United States
- EY017404/EY/NEI NIH HHS/United States
- R01 AI079231/AI/NIAID NIH HHS/United States
- AI57158/AI/NIAID NIH HHS/United States
- CA057973/CA/NCI NIH HHS/United States
- R24 EY017404/EY/NEI NIH HHS/United States
- AI079231/AI/NIAID NIH HHS/United States
- R01 CA057973/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
