Identification of genetic variation on the horse y chromosome and the tracing of male founder lineages in modern breeds
- PMID: 23573227
- PMCID: PMC3616054
- DOI: 10.1371/journal.pone.0060015
Identification of genetic variation on the horse y chromosome and the tracing of male founder lineages in modern breeds
Abstract
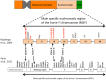
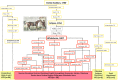
The paternally inherited Y chromosome displays the population genetic history of males. While modern domestic horses (Equus caballus) exhibit abundant diversity within maternally inherited mitochondrial DNA, no significant Y-chromosomal sequence diversity has been detected. We used high throughput sequencing technology to identify the first polymorphic Y-chromosomal markers useful for tracing paternal lines. The nucleotide variability of the modern horse Y chromosome is extremely low, resulting in six haplotypes (HT), all clearly distinct from the Przewalski horse (E. przewalskii). The most widespread HT1 is ancestral and the other five haplotypes apparently arose on the background of HT1 by mutation or gene conversion after domestication. Two haplotypes (HT2 and HT3) are widely distributed at high frequencies among modern European horse breeds. Using pedigree information, we trace the distribution of Y-haplotype diversity to particular founders. The mutation leading to HT3 occurred in the germline of the famous English Thoroughbred stallion "Eclipse" or his son or grandson and its prevalence demonstrates the influence of this popular paternal line on modern sport horse breeds. The pervasive introgression of Thoroughbred stallions during the last 200 years to refine autochthonous breeds has strongly affected the distribution of Y-chromosomal variation in modern horse breeds and has led to the replacement of autochthonous Y chromosomes. Only a few northern European breeds bear unique variants at high frequencies or fixed within but not shared among breeds. Our Y-chromosomal data complement the well established mtDNA lineages and document the male side of the genetic history of modern horse breeds and breeding practices.
Conflict of interest statement
Figures

References
-
- Repping S, van Daalen SK, Brown LG, Korver CM, Lange J, et al. (2006) High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet 38: 463–467. - PubMed
-
- Jobling MA, Samara V, Pandya A, Fretwell N, Bernasconi B, et al. (1996) Recurrent duplication and deletion polymorphisms on the long arm of the Y chromosome in normal males. Hum Mol Genet 5: 1767–1775. - PubMed
-
- Trombetta B, Cruciani F, Underhill PA, Sellitto D, Scozzari R (2009) Footprints of X-to-Y gene conversion in recent human evolution. Mol Biol Evol 27: 714–725. - PubMed
-
- Ellegren H (2011) Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet 12: 157–166. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

