Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells
- PMID: 23573229
- PMCID: PMC3616089
- DOI: 10.1371/journal.pone.0060020
Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells
Erratum in
-
Correction: Utx Is Required for Proper Induction of Ectoderm and Mesoderm during Differentiation of Embryonic Stem Cells.PLoS One. 2024 Jun 27;19(6):e0306360. doi: 10.1371/journal.pone.0306360. eCollection 2024. PLoS One. 2024. PMID: 38935760 Free PMC article.
Abstract
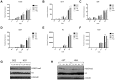
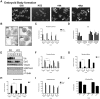
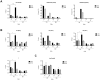
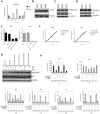
Embryonic development requires chromatin remodeling for dynamic regulation of gene expression patterns to ensure silencing of pluripotent transcription factors and activation of developmental regulators. Demethylation of H3K27me3 by the histone demethylases Utx and Jmjd3 is important for the activation of lineage choice genes in response to developmental signals. To further understand the function of Utx in pluripotency and differentiation we generated Utx knockout embryonic stem cells (ESCs). Here we show that Utx is not required for the proliferation of ESCs, however, Utx contributes to the establishment of ectoderm and mesoderm in vitro. Interestingly, this contribution is independent of the catalytic activity of Utx. Furthermore, we provide data showing that the Utx homologue, Uty, which is devoid of detectable demethylase activity, and Jmjd3 partly compensate for the loss of Utx. Taken together our results show that Utx is required for proper formation of ectoderm and mesoderm in vitro, and that Utx, similar to its C.elegans homologue, has demethylase dependent and independent functions.
Conflict of interest statement
Figures



Similar articles
-
UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity.Proc Natl Acad Sci U S A. 2012 Sep 18;109(38):15324-9. doi: 10.1073/pnas.1204166109. Epub 2012 Sep 4. Proc Natl Acad Sci U S A. 2012. PMID: 22949634 Free PMC article.
-
X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner.Proc Natl Acad Sci U S A. 2012 Aug 7;109(32):13004-9. doi: 10.1073/pnas.1210787109. Epub 2012 Jul 23. Proc Natl Acad Sci U S A. 2012. PMID: 22826230 Free PMC article.
-
KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development.PLoS Genet. 2014 Aug 7;10(8):e1004507. doi: 10.1371/journal.pgen.1004507. eCollection 2014 Aug. PLoS Genet. 2014. PMID: 25101834 Free PMC article.
-
Lysine Demethylase KDM6A in Differentiation, Development, and Cancer.Mol Cell Biol. 2020 Sep 28;40(20):e00341-20. doi: 10.1128/MCB.00341-20. Print 2020 Sep 28. Mol Cell Biol. 2020. PMID: 32817139 Free PMC article. Review.
-
JMJD3: a critical epigenetic regulator in stem cell fate.Cell Commun Signal. 2021 Jul 3;19(1):72. doi: 10.1186/s12964-021-00753-8. Cell Commun Signal. 2021. PMID: 34217316 Free PMC article. Review.
Cited by
-
JMJD3 as an epigenetic regulator in development and disease.Int J Biochem Cell Biol. 2015 Oct;67:148-57. doi: 10.1016/j.biocel.2015.07.006. Epub 2015 Jul 17. Int J Biochem Cell Biol. 2015. PMID: 26193001 Free PMC article. Review.
-
Genome-wide profiling reveals stimulus-specific functions of p53 during differentiation and DNA damage of human embryonic stem cells.Nucleic Acids Res. 2014 Jan;42(1):205-23. doi: 10.1093/nar/gkt866. Epub 2013 Sep 27. Nucleic Acids Res. 2014. PMID: 24078252 Free PMC article.
-
UTX condensation underlies its tumour-suppressive activity.Nature. 2021 Sep;597(7878):726-731. doi: 10.1038/s41586-021-03903-7. Epub 2021 Sep 15. Nature. 2021. PMID: 34526716 Free PMC article.
-
Effect of histone demethylase KDM5B on long-term cognitive impairment in neonatal rats induced by sevoflurane.Front Mol Neurosci. 2024 Nov 27;17:1459358. doi: 10.3389/fnmol.2024.1459358. eCollection 2024. Front Mol Neurosci. 2024. PMID: 39664113 Free PMC article.
-
KDM6A/UTX promotes spermatogenic gene expression across generations and is not required for male fertility†.Biol Reprod. 2024 Feb 10;110(2):391-407. doi: 10.1093/biolre/ioad141. Biol Reprod. 2024. PMID: 37861693 Free PMC article.
References
-
- Niwa H (2007) How is pluripotency determined and maintained? Development 134: 635–646. - PubMed
-
- Evans M (2011) Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat Rev Mol Cell Biol 12: 680–686. - PubMed
-
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599. - PubMed
-
- Lund AH, van Lohuizen M (2004) Epigenetics and cancer. Genes Dev 18: 2315–2335. - PubMed
-
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

