Engineering agatoxin, a cystine-knot peptide from spider venom, as a molecular probe for in vivo tumor imaging
- PMID: 23573262
- PMCID: PMC3616073
- DOI: 10.1371/journal.pone.0060498
Engineering agatoxin, a cystine-knot peptide from spider venom, as a molecular probe for in vivo tumor imaging
Abstract
Background: Cystine-knot miniproteins, also known as knottins, have shown great potential as molecular scaffolds for the development of targeted therapeutics and diagnostic agents. For this purpose, previous protein engineering efforts have focused on knottins based on the Ecballium elaterium trypsin inhibitor (EETI) from squash seeds, the Agouti-related protein (AgRP) neuropeptide from mammals, or the Kalata B1 uterotonic peptide from plants. Here, we demonstrate that Agatoxin (AgTx), an ion channel inhibitor found in spider venom, can be used as a molecular scaffold to engineer knottins that bind with high-affinity to a tumor-associated integrin receptor.
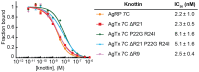
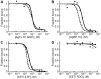
Methodology/principal findings: We used a rational loop-grafting approach to engineer AgTx variants that bound to αvβ3 integrin with affinities in the low nM range. We showed that a disulfide-constrained loop from AgRP, a structurally-related knottin, can be substituted into AgTx to confer its high affinity binding properties. In parallel, we identified amino acid mutations required for efficient in vitro folding of engineered integrin-binding AgTx variants. Molecular imaging was used to evaluate in vivo tumor targeting and biodistribution of an engineered AgTx knottin compared to integrin-binding knottins based on AgRP and EETI. Knottin peptides were chemically synthesized and conjugated to a near-infrared fluorescent dye. Integrin-binding AgTx, AgRP, and EETI knottins all generated high tumor imaging contrast in U87MG glioblastoma xenograft models. Interestingly, EETI-based knottins generated significantly lower non-specific kidney imaging signals compared to AgTx and AgRP-based knottins.
Conclusions/significance: In this study, we demonstrate that AgTx, a knottin from spider venom, can be engineered to bind with high affinity to a tumor-associated receptor target. This work validates AgTx as a viable molecular scaffold for protein engineering, and further demonstrates the promise of using tumor-targeting knottins as probes for in vivo molecular imaging.
Conflict of interest statement
Figures


Similar articles
-
Functional mutation of multiple solvent-exposed loops in the Ecballium elaterium trypsin inhibitor-II cystine knot miniprotein.PLoS One. 2011 Feb 18;6(2):e16112. doi: 10.1371/journal.pone.0016112. PLoS One. 2011. PMID: 21364742 Free PMC article.
-
Evaluation of a (64)Cu-labeled cystine-knot peptide based on agouti-related protein for PET of tumors expressing alphavbeta3 integrin.J Nucl Med. 2010 Feb;51(2):251-258. doi: 10.2967/jnumed.109.069831. J Nucl Med. 2010. PMID: 20124048 Free PMC article.
-
Engineered knottin peptide enables noninvasive optical imaging of intracranial medulloblastoma.Proc Natl Acad Sci U S A. 2013 Sep 3;110(36):14598-603. doi: 10.1073/pnas.1311333110. Epub 2013 Aug 15. Proc Natl Acad Sci U S A. 2013. PMID: 23950221 Free PMC article.
-
Cy5.5-Knottin 2.5F.2009 Mar 26 [updated 2009 May 28]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. 2009 Mar 26 [updated 2009 May 28]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. PMID: 20641522 Free Books & Documents. Review.
-
Cy5.5-Knottin 2.5D.2009 Mar 26 [updated 2009 May 28]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. 2009 Mar 26 [updated 2009 May 28]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. PMID: 20641277 Free Books & Documents. Review.
Cited by
-
Utilizing combinatorial engineering to develop Tie2 targeting antagonistic angiopoetin-2 ligands as candidates for anti-angiogenesis therapy.Oncotarget. 2017 May 16;8(20):33571-33585. doi: 10.18632/oncotarget.16827. Oncotarget. 2017. PMID: 28422724 Free PMC article.
-
A workflow for in silico design of hIL-10 and ebvIL-10 inhibitors using well-known miniprotein scaffolds.J Mol Model. 2017 Apr;23(4):118. doi: 10.1007/s00894-017-3276-1. Epub 2017 Mar 14. J Mol Model. 2017. PMID: 28293795
-
Applications of Yeast Surface Display for Protein Engineering.Methods Mol Biol. 2015;1319:155-75. doi: 10.1007/978-1-4939-2748-7_8. Methods Mol Biol. 2015. PMID: 26060074 Free PMC article. Review.
-
CNS Anticancer Drug Discovery and Development Conference White Paper.Neuro Oncol. 2015 Nov;17 Suppl 6(Suppl 6):vi1-26. doi: 10.1093/neuonc/nov169. Neuro Oncol. 2015. PMID: 26403167 Free PMC article.
-
Molecular evolution of peptides by yeast surface display technology.Medchemcomm. 2019 Jul 10;10(9):1569-1580. doi: 10.1039/c9md00252a. eCollection 2019 Sep 1. Medchemcomm. 2019. PMID: 31803399 Free PMC article. Review.
References
-
- Friedman M, Stahl S (2009) Engineered affinity proteins for tumour-targeting applications. Biotechnol Appl Biochem 53: 1–29. - PubMed
-
- Batra SK, Jain M, Wittel UA, Chauhan SC, Colcher D (2002) Pharmacokinetics and biodistribution of genetically engineered antibodies. Curr Opin Biotechnol 13: 603–608. - PubMed
-
- Daly NL, Craik DJ (2011) Bioactive cystine knot proteins. Curr Opin Chem Biol 15: 362–368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

