Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays
- PMID: 23573916
- PMCID: PMC4834911
- DOI: 10.1111/gtc.12050
Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays
Abstract
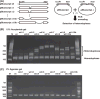
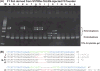
The heteroduplex mobility assay (HMA) is widely used to characterize strain variants of human viruses. To determine whether it can detect small sequence differences in homologous templates, we constructed a series of deletion constructs (1-10 bp deletions) in the multiple cloning site (MCS) of pBluescript II. After PCR amplification of the MCS using a mixture of wild-type and one of the deletion constructs, the resulting PCR amplicons were electrophoresed using 15% polyacrylamide gels. Two types of heteroduplexes exhibited retarded electrophoretic migration compared with individual homoduplexes. Therefore, we applied this HMA to detect transcription activator-like effector nucleases (TALEN)-induced insertion and/or deletion (indel) mutations at an endogenous locus. We found that TALEN in vivo activity was easily estimated by the degree of multiple HMA profiles derived from TALEN-injected F0 embryos. Furthermore, TALEN-injected F0 founder fish produced several unique HMA profiles in F1 embryos. Sequence analysis confirmed that the different HMA profiles contained distinct indel mutations. Thus, HMA is a rapid and sensitive analytical method for the detection of the TALEN-mediated genome modifications.
© 2013 The Authors Genes to Cells © 2013 by the Molecular Biology Society of Japan and Wiley Publishing Asia Pty Ltd.
Figures



References
-
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

