LXCXE-independent chromatin remodeling by Rb/E2f mediates neuronal quiescence
- PMID: 23574720
- PMCID: PMC3674069
- DOI: 10.4161/cc.24527
LXCXE-independent chromatin remodeling by Rb/E2f mediates neuronal quiescence
Abstract
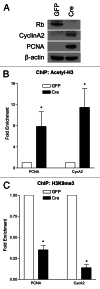
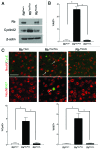
Neuronal survival is dependent upon the retinoblastoma family members, Rb1 (Rb) and Rb2 (p130). Rb is thought to regulate gene repression, in part, through direct recruitment of chromatin modifying enzymes to its conserved LXCXE binding domain. We sought to examine the mechanisms that Rb employs to mediate cell cycle gene repression in terminally differentiated cortical neurons. Here, we report that Rb loss converts chromatin at the promoters of E2f-target genes to an activated state. We established a mouse model system in which Rb-LXCXE interactions could be induciblely disabled. Surprisingly, this had no effect on survival or gene silencing in neuronal quiescence. Absence of the Rb LXCXE-binding domain in neurons is compatible with gene repression and long-term survival, unlike Rb deficiency. Finally, we are able to show that chromatin activation following Rb deletion occurs at the level of E2fs. Blocking E2f-mediated transcription downstream of Rb loss is sufficient to maintain chromatin in an inactive state. Taken together our results suggest a model whereby Rb-E2f interactions are sufficient to maintain gene repression irrespective of LXCXE-dependent chromatin remodeling.
Keywords: E2f; Rb; cell cycle; neuronal quiescence; transcription.
Figures


Comment in
-
Rb and chromatin remodeling in the maintenance of the post-mitotic state of neurons.Cell Cycle. 2013 Jun 1;12(11):1661-2. doi: 10.4161/cc.25071. Epub 2013 May 17. Cell Cycle. 2013. PMID: 23708446 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
