M-CSF instructs myeloid lineage fate in single haematopoietic stem cells
- PMID: 23575636
- PMCID: PMC3679883
- DOI: 10.1038/nature12026
M-CSF instructs myeloid lineage fate in single haematopoietic stem cells
Abstract
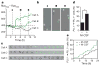
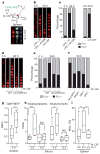
Under stress conditions such as infection or inflammation the body rapidly needs to generate new blood cells that are adapted to the challenge. Haematopoietic cytokines are known to increase output of specific mature cells by affecting survival, expansion and differentiation of lineage-committed progenitors, but it has been debated whether long-term haematopoietic stem cells (HSCs) are susceptible to direct lineage-specifying effects of cytokines. Although genetic changes in transcription factor balance can sensitize HSCs to cytokine instruction, the initiation of HSC commitment is generally thought to be triggered by stochastic fluctuation in cell-intrinsic regulators such as lineage-specific transcription factors, leaving cytokines to ensure survival and proliferation of the progeny cells. Here we show that macrophage colony-stimulating factor (M-CSF, also called CSF1), a myeloid cytokine released during infection and inflammation, can directly induce the myeloid master regulator PU.1 and instruct myeloid cell-fate change in mouse HSCs, independently of selective survival or proliferation. Video imaging and single-cell gene expression analysis revealed that stimulation of highly purified HSCs with M-CSF in culture resulted in activation of the PU.1 promoter and an increased number of PU.1(+) cells with myeloid gene signature and differentiation potential. In vivo, high systemic levels of M-CSF directly stimulated M-CSF-receptor-dependent activation of endogenous PU.1 protein in single HSCs and induced a PU.1-dependent myeloid differentiation preference. Our data demonstrate that lineage-specific cytokines can act directly on HSCs in vitro and in vivo to instruct a change of cell identity. This fundamentally changes the current view of how HSCs respond to environmental challenge and implicates stress-induced cytokines as direct instructors of HSC fate.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Comment in
-
Haematopoiesis: early decisions.Nat Rev Immunol. 2013 May;13(5):304. doi: 10.1038/nri3454. Nat Rev Immunol. 2013. PMID: 23618825 No abstract available.
-
Lineage specification: reading the instructions may help!Curr Biol. 2013 Aug 5;23(15):R662-5. doi: 10.1016/j.cub.2013.06.054. Curr Biol. 2013. PMID: 23928087
References
-
- Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC, Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325:217–218. - PubMed
-
- Sarrazin S, et al. MafB restricts M-CSF dependent myeloid commitment divisions of hematopoietic stem cells. Cell. 2009;138:300–313. - PubMed
-
- Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nature Rev Genet. 2000;1:57–64. - PubMed
-
- Cantor AB, Orkin SH. Hematopoietic development: a balancing act. Curr Opin Genet Dev. 2001;11:513–519. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

