Rapid evolution of Beta-keratin genes contribute to phenotypic differences that distinguish turtles and birds from other reptiles
- PMID: 23576313
- PMCID: PMC3673632
- DOI: 10.1093/gbe/evt060
Rapid evolution of Beta-keratin genes contribute to phenotypic differences that distinguish turtles and birds from other reptiles
Abstract
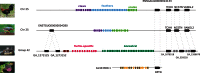
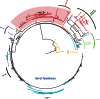
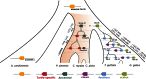
Sequencing of vertebrate genomes permits changes in distinct protein families, including gene gains and losses, to be ascribed to lineage-specific phenotypes. A prominent example of this is the large-scale duplication of beta-keratin genes in the ancestors of birds, which was crucial to the subsequent evolution of their beaks, claws, and feathers. Evidence suggests that the shell of Pseudomys nelsoni contains at least 16 beta-keratins proteins, but it is unknown whether this is a complete set and whether their corresponding genes are orthologous to avian beak, claw, or feather beta-keratin genes. To address these issues and to better understand the evolution of the turtle shell at a molecular level, we surveyed the diversity of beta-keratin genes from the genome assemblies of three turtles, Chrysemys picta, Pelodiscus sinensis, and Chelonia mydas, which together represent over 160 Myr of chelonian evolution. For these three turtles, we found 200 beta-keratins, which indicate that, as for birds, a large expansion of beta-keratin genes in turtles occurred concomitantly with the evolution of a unique phenotype, namely, their plastron and carapace. Phylogenetic reconstruction of beta-keratin gene evolution suggests that separate waves of gene duplication within a single genomic location gave rise to scales, claws, and feathers in birds, and independently the scutes of the shell in turtles.
Keywords: Sauropsids; beta-keratins; gene duplication; turtle shell.
Figures

Similar articles
-
Dynamic evolution of the alpha (α) and beta (β) keratins has accompanied integument diversification and the adaptation of birds into novel lifestyles.BMC Evol Biol. 2014 Dec 12;14:249. doi: 10.1186/s12862-014-0249-1. BMC Evol Biol. 2014. PMID: 25496280 Free PMC article.
-
Review: Evolution and diversification of corneous beta-proteins, the characteristic epidermal proteins of reptiles and birds.J Exp Zool B Mol Dev Evol. 2018 Dec;330(8):438-453. doi: 10.1002/jez.b.22840. Epub 2019 Jan 14. J Exp Zool B Mol Dev Evol. 2018. PMID: 30637919 Review.
-
Genomic organization and molecular phylogenies of the beta (beta) keratin multigene family in the chicken (Gallus gallus) and zebra finch (Taeniopygia guttata): implications for feather evolution.BMC Evol Biol. 2010 May 18;10:148. doi: 10.1186/1471-2148-10-148. BMC Evol Biol. 2010. PMID: 20482795 Free PMC article.
-
Molecular evolution and expression of archosaurian β-keratins: diversification and expansion of archosaurian β-keratins and the origin of feather β-keratins.J Exp Zool B Mol Dev Evol. 2013 Sep;320(6):393-405. doi: 10.1002/jez.b.22514. Epub 2013 Jun 6. J Exp Zool B Mol Dev Evol. 2013. PMID: 23744807
-
Sauropsids Cornification is Based on Corneous Beta-Proteins, a Special Type of Keratin-Associated Corneous Proteins of the Epidermis.J Exp Zool B Mol Dev Evol. 2016 Sep;326(6):338-351. doi: 10.1002/jez.b.22689. Epub 2016 Aug 10. J Exp Zool B Mol Dev Evol. 2016. PMID: 27506161 Review.
Cited by
-
Genomic organization, transcriptomic analysis, and functional characterization of avian α- and β-keratins in diverse feather forms.Genome Biol Evol. 2014 Aug 24;6(9):2258-73. doi: 10.1093/gbe/evu181. Genome Biol Evol. 2014. PMID: 25152353 Free PMC article.
-
Two Antarctic penguin genomes reveal insights into their evolutionary history and molecular changes related to the Antarctic environment.Gigascience. 2014 Dec 12;3(1):27. doi: 10.1186/2047-217X-3-27. eCollection 2014. Gigascience. 2014. PMID: 25671092 Free PMC article.
-
Development-Associated Genes of the Epidermal Differentiation Complex (EDC).J Dev Biol. 2024 Jan 15;12(1):4. doi: 10.3390/jdb12010004. J Dev Biol. 2024. PMID: 38248869 Free PMC article. Review.
-
Dynamic evolution of the alpha (α) and beta (β) keratins has accompanied integument diversification and the adaptation of birds into novel lifestyles.BMC Evol Biol. 2014 Dec 12;14:249. doi: 10.1186/s12862-014-0249-1. BMC Evol Biol. 2014. PMID: 25496280 Free PMC article.
-
Insights into the adaptive evolution of chromosome and essential traits through chromosome-level genome assembly of Gekko japonicus.iScience. 2023 Nov 14;27(1):108445. doi: 10.1016/j.isci.2023.108445. eCollection 2024 Jan 19. iScience. 2023. PMID: 38205241 Free PMC article.
References
-
- Alibardi L. Immunocytochemical observations on the cornification of soft and hard epidermis in the turtle Chrysemys picta. Zoology (Jena) 2002;105(1):31–44. - PubMed
-
- Alibardi L, Toni M, Dalla Valle L. Hard cornification in reptilian epidermis in comparison to cornification in mammalian epidermis. Exp Dermatol. 2007;16:961–976. - PubMed
-
- Barten R, Torkar M, Haude A, Trowsdale J, Wilson MJ. Divergent and convergent evolution of nk-cell receptors. Trends Immunol. 2001;22(1):52–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

