Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids
- PMID: 23576513
- PMCID: PMC3676120
- DOI: 10.1128/JVI.00582-13
Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids
Abstract
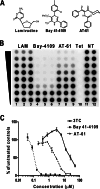
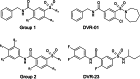
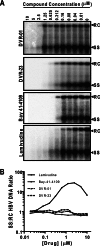
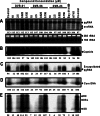
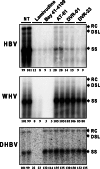
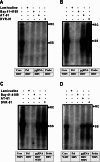
Chronic hepatitis B virus (HBV) infection, a serious public health problem leading to cirrhosis and hepatocellular carcinoma, is currently treated with either pegylated alpha interferon (pegIFN-α) or one of the five nucleos(t)ide analogue viral DNA polymerase inhibitors. However, neither pegIFN-α nor nucleos(t)ide analogues are capable of reliably curing the viral infection. In order to develop novel antiviral drugs against HBV, we established a cell-based screening assay by using an immortalized mouse hepatocyte-derived stable cell line supporting a high level of HBV replication in a tetracycline-inducible manner. Screening of a library consisting of 26,900 small molecules led to the discovery of a series of sulfamoylbenzamide (SBA) derivatives that significantly reduced the amount of cytoplasmic HBV DNA. Structure-activity relationship studies have thus far identified a group of fluorine-substituted SBAs with submicromolar antiviral activity against HBV in human hepatoma cells. Mechanistic analyses reveal that the compounds dose dependently inhibit the formation of pregenomic RNA (pgRNA)-containing nucleocapsids of HBV but not other animal hepadnaviruses, such as woodchuck hepatitis virus (WHV) and duck hepatitis B virus (DHBV). Moreover, heterologous genetic complementation studies of capsid protein, DNA polymerase, and pgRNA between HBV and WHV suggest that HBV capsid protein confers sensitivity to the SBAs. In summary, SBAs represent a novel chemical entity with superior activity and a unique antiviral mechanism and are thus warranted for further development as novel antiviral therapeutics for the treatment of chronic hepatitis B.
Figures
Similar articles
-
Identification and characterization of a novel hepatitis B virus pregenomic RNA encapsidation inhibitor.Antiviral Res. 2020 Mar;175:104709. doi: 10.1016/j.antiviral.2020.104709. Epub 2020 Jan 12. Antiviral Res. 2020. PMID: 31940474
-
Discovery and Mechanistic Study of Benzamide Derivatives That Modulate Hepatitis B Virus Capsid Assembly.J Virol. 2017 Jul 27;91(16):e00519-17. doi: 10.1128/JVI.00519-17. Print 2017 Aug 15. J Virol. 2017. PMID: 28566379 Free PMC article.
-
Hepatitis B Virus Capsid Assembly Modulators, but Not Nucleoside Analogs, Inhibit the Production of Extracellular Pregenomic RNA and Spliced RNA Variants.Antimicrob Agents Chemother. 2017 Jul 25;61(8):e00680-17. doi: 10.1128/AAC.00680-17. Print 2017 Aug. Antimicrob Agents Chemother. 2017. PMID: 28559265 Free PMC article.
-
Targeting the multifunctional HBV core protein as a potential cure for chronic hepatitis B.Antiviral Res. 2020 Oct;182:104917. doi: 10.1016/j.antiviral.2020.104917. Epub 2020 Aug 17. Antiviral Res. 2020. PMID: 32818519 Free PMC article. Review.
-
HBV Genome and Life Cycle.Adv Exp Med Biol. 2020;1179:17-37. doi: 10.1007/978-981-13-9151-4_2. Adv Exp Med Biol. 2020. PMID: 31741332 Review.
Cited by
-
Core protein: A pleiotropic keystone in the HBV lifecycle.Antiviral Res. 2015 Sep;121:82-93. doi: 10.1016/j.antiviral.2015.06.020. Epub 2015 Jun 27. Antiviral Res. 2015. PMID: 26129969 Free PMC article. Review.
-
CpAMs induce assembly of HBV capsids with altered electrophoresis mobility: Implications for mechanism of inhibiting pgRNA packaging.Antiviral Res. 2018 Nov;159:1-12. doi: 10.1016/j.antiviral.2018.09.001. Epub 2018 Sep 7. Antiviral Res. 2018. PMID: 30201396 Free PMC article.
-
Novel Hepatitis B Virus Capsid-Targeting Antiviral That Aggregates Core Particles and Inhibits Nuclear Entry of Viral Cores.ACS Infect Dis. 2019 May 10;5(5):750-758. doi: 10.1021/acsinfecdis.8b00235. Epub 2019 Jan 14. ACS Infect Dis. 2019. PMID: 30582687 Free PMC article.
-
HBV cccDNA-A Culprit and Stumbling Block for the Hepatitis B Virus Infection: Its Presence in Hepatocytes Perplexed the Possible Mission for a Functional Cure.ACS Omega. 2022 Jul 7;7(28):24066-24081. doi: 10.1021/acsomega.2c02216. eCollection 2022 Jul 19. ACS Omega. 2022. PMID: 35874215 Free PMC article. Review.
-
Constrained evolution of overlapping genes in viral host adaptation: Acquisition of glycosylation motifs in hepadnaviral precore/core genes.PLoS Pathog. 2022 Jul 28;18(7):e1010739. doi: 10.1371/journal.ppat.1010739. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35901192 Free PMC article.
References
-
- McMahon BJ. 2005. Epidemiology and natural history of hepatitis B. Semin. Liver Dis. 25(Suppl 1):3–8 - PubMed
-
- Lee WM. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733–1745 - PubMed
-
- Lok AS. 2004. Prevention of hepatitis B virus-related hepatocellular carcinoma. Gastroenterology 127:S303–309 - PubMed
-
- Keeffe EB, Dieterich DT, Han SH, Jacobson IM, Martin P, Schiff ER, Tobias H. 2008. A treatment algorithm for the management of chronic hepatitis B virus infection in the United States: 2008 update. Clin. Gastroenterol. Hepatol. 6:1315–1341 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

