PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains
- PMID: 23576556
- PMCID: PMC3673830
- DOI: 10.1158/0008-5472.CAN-12-3292
PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains
Abstract
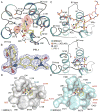
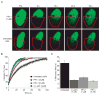
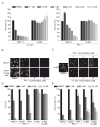
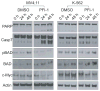
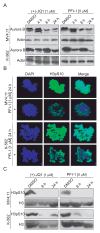
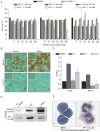
Bromo and extra terminal (BET) proteins (BRD2, BRD3, BRD4, and BRDT) are transcriptional regulators required for efficient expression of several growth promoting and antiapoptotic genes as well as for cell-cycle progression. BET proteins are recruited on transcriptionally active chromatin via their two N-terminal bromodomains (BRD), a protein interaction module that specifically recognizes acetylated lysine residues in histones H3 and H4. Inhibition of the BET-histone interaction results in transcriptional downregulation of a number of oncogenes, providing a novel pharmacologic strategy for the treatment of cancer. Here, we present a potent and highly selective dihydroquinazoline-2-one inhibitor, PFI-1, which efficiently blocks the interaction of BET BRDs with acetylated histone tails. Cocrystal structures showed that PFI-1 acts as an acetyl-lysine (Kac) mimetic inhibitor efficiently occupying the Kac binding site in BRD4 and BRD2. PFI-1 has antiproliferative effects on leukemic cell lines and efficiently abrogates their clonogenic growth. Exposure of sensitive cell lines with PFI-1 results in G1 cell-cycle arrest, downregulation of MYC expression, as well as induction of apoptosis and induces differentiation of primary leukemic blasts. Intriguingly, cells exposed to PFI-1 showed significant downregulation of Aurora B kinase, thus attenuating phosphorylation of the Aurora substrate H3S10, providing an alternative strategy for the specific inhibition of this well-established oncology target.
©2013 AACR.
Conflict of interest statement
Figures

Similar articles
-
Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery.Oncotarget. 2015 Mar 20;6(8):5501-16. doi: 10.18632/oncotarget.3551. Oncotarget. 2015. PMID: 25849938 Free PMC article. Review.
-
Metabolically Derived Lysine Acylations and Neighboring Modifications Tune the Binding of the BET Bromodomains to Histone H4.Biochemistry. 2017 Oct 17;56(41):5485-5495. doi: 10.1021/acs.biochem.7b00595. Epub 2017 Oct 5. Biochemistry. 2017. PMID: 28945351 Free PMC article.
-
AZD5153: A Novel Bivalent BET Bromodomain Inhibitor Highly Active against Hematologic Malignancies.Mol Cancer Ther. 2016 Nov;15(11):2563-2574. doi: 10.1158/1535-7163.MCT-16-0141. Epub 2016 Aug 29. Mol Cancer Ther. 2016. PMID: 27573426
-
Affinity map of bromodomain protein 4 (BRD4) interactions with the histone H4 tail and the small molecule inhibitor JQ1.J Biol Chem. 2014 Mar 28;289(13):9304-19. doi: 10.1074/jbc.M113.523019. Epub 2014 Feb 4. J Biol Chem. 2014. PMID: 24497639 Free PMC article.
-
Small-Molecule Targeting of BET Proteins in Cancer.Adv Cancer Res. 2016;131:21-58. doi: 10.1016/bs.acr.2016.04.001. Epub 2016 May 31. Adv Cancer Res. 2016. PMID: 27451123 Review.
Cited by
-
Combining Data-Driven and Structure-Based Approaches in Designing Dual PARP1-BRD4 Inhibitors for Breast Cancer Treatment.J Chem Inf Model. 2024 Oct 14;64(19):7725-7742. doi: 10.1021/acs.jcim.4c01421. Epub 2024 Sep 18. J Chem Inf Model. 2024. PMID: 39292752 Free PMC article.
-
Protein Acetylation at the Interface of Genetics, Epigenetics and Environment in Cancer.Metabolites. 2021 Apr 1;11(4):216. doi: 10.3390/metabo11040216. Metabolites. 2021. PMID: 33916219 Free PMC article. Review.
-
Roles of Bromodomain Extra Terminal Proteins in Metabolic Signaling and Diseases.Pharmaceuticals (Basel). 2022 Aug 22;15(8):1032. doi: 10.3390/ph15081032. Pharmaceuticals (Basel). 2022. PMID: 36015180 Free PMC article. Review.
-
Privileged diazepine compounds and their emergence as bromodomain inhibitors.Chem Biol. 2014 May 22;21(5):573-83. doi: 10.1016/j.chembiol.2014.03.004. Epub 2014 Apr 17. Chem Biol. 2014. PMID: 24746559 Free PMC article. Review.
-
Synthesis and biological evaluation of N-(3-oxo-3,4-dihydro-2H-benzo[b][1,4]oxazin-7-yl)benzenesulfonamide derivatives as new BET bromodomain inhibitors for anti-hematologic malignancies activities.Mol Divers. 2017 Feb;21(1):125-136. doi: 10.1007/s11030-016-9707-6. Epub 2016 Nov 17. Mol Divers. 2017. PMID: 27858214
References
-
- Mujtaba S, He Y, Zeng L, Farooq A, Carlson JE, Ott M, et al. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol Cell. 2002;9:575–86. - PubMed
-
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

