Probing DNA shape and methylation state on a genomic scale with DNase I
- PMID: 23576721
- PMCID: PMC3631675
- DOI: 10.1073/pnas.1216822110
Probing DNA shape and methylation state on a genomic scale with DNase I
Abstract
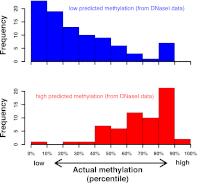
DNA binding proteins find their cognate sequences within genomic DNA through recognition of specific chemical and structural features. Here we demonstrate that high-resolution DNase I cleavage profiles can provide detailed information about the shape and chemical modification status of genomic DNA. Analyzing millions of DNA backbone hydrolysis events on naked genomic DNA, we show that the intrinsic rate of cleavage by DNase I closely tracks the width of the minor groove. Integration of these DNase I cleavage data with bisulfite sequencing data for the same cell type's genome reveals that cleavage directly adjacent to cytosine-phosphate-guanine (CpG) dinucleotides is enhanced at least eightfold by cytosine methylation. This phenomenon we show to be attributable to methylation-induced narrowing of the minor groove. Furthermore, we demonstrate that it enables simultaneous mapping of DNase I hypersensitivity and regional DNA methylation levels using dense in vivo cleavage data. Taken together, our results suggest a general mechanism by which CpG methylation can modulate protein-DNA interaction strength via the remodeling of DNA shape.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Systematic prediction of DNA shape changes due to CpG methylation explains epigenetic effects on protein-DNA binding.Epigenetics Chromatin. 2018 Feb 6;11(1):6. doi: 10.1186/s13072-018-0174-4. Epigenetics Chromatin. 2018. PMID: 29409522 Free PMC article.
-
Large-scale methylation analysis of human genomic DNA reveals tissue-specific differences between the methylation profiles of genes and pseudogenes.Hum Mol Genet. 2000 Nov 1;9(18):2651-63. doi: 10.1093/hmg/9.18.2651. Hum Mol Genet. 2000. PMID: 11063724
-
Predicting genome-wide DNA methylation using methylation marks, genomic position, and DNA regulatory elements.Genome Biol. 2015 Jan 24;16(1):14. doi: 10.1186/s13059-015-0581-9. Genome Biol. 2015. PMID: 25616342 Free PMC article.
-
DNA recognition by DNase I.J Mol Recognit. 1994 Jun;7(2):65-70. doi: 10.1002/jmr.300070203. J Mol Recognit. 1994. PMID: 7826675 Review.
-
Study of tissue-specific CpG methylation of DNA in extended genomic loci.Biochemistry (Mosc). 2005 May;70(5):596-603. doi: 10.1007/s10541-005-0153-9. Biochemistry (Mosc). 2005. PMID: 15948713 Review.
Cited by
-
Global reference mapping of human transcription factor footprints.Nature. 2020 Jul;583(7818):729-736. doi: 10.1038/s41586-020-2528-x. Epub 2020 Jul 29. Nature. 2020. PMID: 32728250 Free PMC article.
-
Systematic prediction of DNA shape changes due to CpG methylation explains epigenetic effects on protein-DNA binding.Epigenetics Chromatin. 2018 Feb 6;11(1):6. doi: 10.1186/s13072-018-0174-4. Epigenetics Chromatin. 2018. PMID: 29409522 Free PMC article.
-
Charting epimutation dynamics in human hematopoietic differentiation.Blood Sci. 2024 Jun 13;6(3):e00197. doi: 10.1097/BS9.0000000000000197. eCollection 2024 Jul. Blood Sci. 2024. PMID: 38872911 Free PMC article.
-
Evolving insights on how cytosine methylation affects protein-DNA binding.Brief Funct Genomics. 2015 Jan;14(1):61-73. doi: 10.1093/bfgp/elu040. Epub 2014 Oct 14. Brief Funct Genomics. 2015. PMID: 25319759 Free PMC article. Review.
-
Conservation of trans-acting circuitry during mammalian regulatory evolution.Nature. 2014 Nov 20;515(7527):365-70. doi: 10.1038/nature13972. Nature. 2014. PMID: 25409825 Free PMC article.
References
-
- Oliveri M, et al. DNase I behaves as a transcription factor which modulates Fas expression in human cells. Eur J Immunol. 2004;34(1):273–279. - PubMed
-
- Suck D, Lahm A, Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988;332(6163):464–468. - PubMed
-
- Weston SA, Lahm A, Suck D. X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution. J Mol Biol. 1992;226(4):1237–1256. - PubMed
-
- Heddi B, Abi-Ghanem J, Lavigne M, Hartmann B. Sequence-dependent DNA flexibility mediates DNase I cleavage. J Mol Biol. 2010;395(1):123–133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

