Transitions to catalytically inactive conformations in EGFR kinase
- PMID: 23576739
- PMCID: PMC3645566
- DOI: 10.1073/pnas.1220843110
Transitions to catalytically inactive conformations in EGFR kinase
Abstract
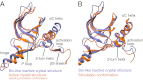
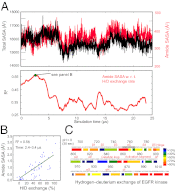
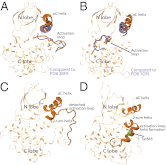
The epidermal growth factor receptor (EGFR) is a key protein in cellular signaling, and its kinase domain (EGFR kinase) is an intensely pursued target of small-molecule drugs. Although both catalytically active and inactive conformations of EGFR kinase have been resolved crystallographically, experimental characterization of the transitions between these conformations remains difficult. Using unbiased, all-atom molecular dynamics simulations, we observed EGFR kinase spontaneously transition from the active to the so-called "Src-like inactive" conformation by way of two sets of intermediate conformations: One corresponds to a previously identified locally disordered state and the other to previously undescribed "extended" conformations, marked by the opening of the ATP-binding site between the two lobes of the kinase domain. We also simulated the protonation-dependent transition of EGFR kinase to another ["Asp-Phe-Gly-out" ("DFG-out")] inactive conformation and observed similar intermediate conformations. A key element observed in the simulated transitions is local unfolding, or "cracking," which supports a prediction of energy landscape theory. We used hydrogen-deuterium (H/D) exchange measurements to corroborate our simulations and found that the simulated intermediate conformations correlate better with the H/D exchange data than existing active or inactive EGFR kinase crystal structures. The intermediate conformations revealed by our simulations of the transition process differ significantly from the existing crystal structures and may provide unique possibilities for structure-based drug discovery.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Disorder guides protein function.Proc Natl Acad Sci U S A. 2013 Apr 30;110(18):7114-5. doi: 10.1073/pnas.1305236110. Epub 2013 Apr 22. Proc Natl Acad Sci U S A. 2013. PMID: 23610426 Free PMC article. No abstract available.
References
-
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–137. - PubMed
-
- Lurje G, Lenz H-J. EGFR signaling and drug discovery. Oncology. 2009;77(6):400–410. - PubMed
-
- Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277(48):46265–46272. - PubMed
-
- Wood ER, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): Relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64(18):6652–6659. - PubMed
-
- Zhang XW, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125(6):1137–1149. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

