Imaging burst kinetics and spatial coordination during serial killing by single natural killer cells
- PMID: 23576740
- PMCID: PMC3631668
- DOI: 10.1073/pnas.1221312110
Imaging burst kinetics and spatial coordination during serial killing by single natural killer cells
Abstract
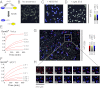
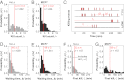
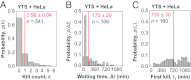
Cytotoxic lymphocytes eliminate virus-infected and cancerous cells by immune recognition and killing through the perforin-granzyme pathway. Traditional killing assays measure average target cell lysis at fixed times and high effector:target ratios. Such assays obscure kinetic details that might reveal novel physiology. We engineered target cells to report on granzyme activity, used very low effector:target ratios to observe potential serial killing, and performed low magnification time-lapse imaging to reveal time-dependent statistics of natural killer (NK) killing at the single-cell level. Most kills occurred during serial killing, and a single NK cell killed up to 10 targets over a 6-h assay. The first kill was slower than subsequent kills, especially on poor targets, or when NK signaling pathways were partially inhibited. Spatial analysis showed that sequential kills were usually adjacent. We propose that NK cells integrate signals from the previous and current target, possibly by simultaneous contact. The resulting burst kinetics and spatial coordination may control the activity of NK cells in tissues.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: From immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–998. - PubMed
-
- Grakoui A, et al. The immunological synapse: A molecular machine controlling T cell activation. Science. 1999;285(5425):221–227. - PubMed
-
- Zagury D, Bernard J, Thierness N, Feldman M, Berke G. Isolation and characterization of individual functionally reactive cytotoxic T lymphocytes: Conjugation, killing and recycling at the single cell level. Eur J Immunol. 1975;5(12):818–822.
-
- Sanderson CJ. The mechanism of T cell mediated cytotoxicity. II. Morphological studies of cell death by time-lapse microcinematography. Proc R Soc Lond B Biol Sci. 1976;192(1107):241–255. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

