Dynein and dynactin leverage their bivalent character to form a high-affinity interaction
- PMID: 23577064
- PMCID: PMC3618186
- DOI: 10.1371/journal.pone.0059453
Dynein and dynactin leverage their bivalent character to form a high-affinity interaction
Erratum in
-
Correction: Dynein and Dynactin Leverage Their Bivalent Character to Form a High-Affinity Interaction.PLoS One. 2024 Jun 4;19(6):e0304916. doi: 10.1371/journal.pone.0304916. eCollection 2024. PLoS One. 2024. PMID: 38833489 Free PMC article.
Abstract
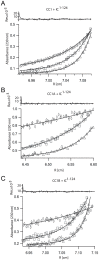
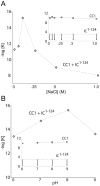
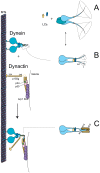
Cytoplasmic dynein and dynactin participate in retrograde transport of organelles, checkpoint signaling and cell division. The principal subunits that mediate this interaction are the dynein intermediate chain (IC) and the dynactin p150(Glued); however, the interface and mechanism that regulates this interaction remains poorly defined. Herein, we use multiple methods to show the N-terminus of mammalian dynein IC, residues 10-44, is sufficient for binding p150(Glued). Consistent with this mapping, monoclonal antibodies that antagonize the dynein-dynactin interaction also bind to this region of the IC. Furthermore, double and triple alanine point mutations spanning residues 6 to 19 in the yeast IC homolog, Pac11, produce significant defects in spindle positioning. Using the same methods we show residues 381 to 530 of p150(Glued) form a minimal fragment that binds to the dynein IC. Sedimentation equilibrium experiments indicate that these individual fragments are predominantly monomeric, but admixtures of the IC and p150(Glued) fragments produce a 2:2 complex. This tetrameric complex is sensitive to salt, temperature and pH, suggesting that the binding is dominated by electrostatic interactions. Finally, circular dichroism (CD) experiments indicate that the N-terminus of the IC is disordered and becomes ordered upon binding p150(Glued). Taken together, the data indicate that the dynein-dynactin interaction proceeds through a disorder-to-order transition, leveraging its bivalent-bivalent character to form a high affinity, but readily reversible interaction.
Conflict of interest statement
Figures






Similar articles
-
Multivalency, autoinhibition, and protein disorder in the regulation of interactions of dynein intermediate chain with dynactin and the nuclear distribution protein.Elife. 2022 Nov 23;11:e80217. doi: 10.7554/eLife.80217. Elife. 2022. PMID: 36416224 Free PMC article.
-
Intrinsic disorder in dynein intermediate chain modulates its interactions with NudE and dynactin.J Biol Chem. 2012 Jul 20;287(30):24884-93. doi: 10.1074/jbc.M112.376038. Epub 2012 Jun 5. J Biol Chem. 2012. PMID: 22669947 Free PMC article.
-
Apoptotic cleavage of cytoplasmic dynein intermediate chain and p150(Glued) stops dynein-dependent membrane motility.J Cell Biol. 2001 Jun 25;153(7):1415-26. doi: 10.1083/jcb.153.7.1415. J Cell Biol. 2001. PMID: 11425872 Free PMC article.
-
Structural dynamics and multiregion interactions in dynein-dynactin recognition.J Biol Chem. 2011 Nov 11;286(45):39349-59. doi: 10.1074/jbc.M111.296277. Epub 2011 Sep 19. J Biol Chem. 2011. PMID: 21931160 Free PMC article.
-
Interplay of Disorder and Sequence Specificity in the Formation of Stable Dynein-Dynactin Complexes.Biophys J. 2020 Sep 1;119(5):950-965. doi: 10.1016/j.bpj.2020.07.023. Epub 2020 Aug 5. Biophys J. 2020. PMID: 32814057 Free PMC article.
Cited by
-
Inositol hexakisphosphate kinase 1 (IP6K1) activity is required for cytoplasmic dynein-driven transport.Biochem J. 2016 Oct 1;473(19):3031-47. doi: 10.1042/BCJ20160610. Epub 2016 Jul 29. Biochem J. 2016. PMID: 27474409 Free PMC article.
-
Multivalency, autoinhibition, and protein disorder in the regulation of interactions of dynein intermediate chain with dynactin and the nuclear distribution protein.Elife. 2022 Nov 23;11:e80217. doi: 10.7554/eLife.80217. Elife. 2022. PMID: 36416224 Free PMC article.
-
The Generation of Dynein Networks by Multi-Layered Regulation and Their Implication in Cell Division.Front Cell Dev Biol. 2020 Jan 31;8:22. doi: 10.3389/fcell.2020.00022. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32083077 Free PMC article. Review.
-
Recruitment of two dyneins to an mRNA-dependent Bicaudal D transport complex.Elife. 2018 Jun 26;7:e36306. doi: 10.7554/eLife.36306. Elife. 2018. PMID: 29944116 Free PMC article.
-
Integrated regulation of motor-driven organelle transport by scaffolding proteins.Trends Cell Biol. 2014 Oct;24(10):564-74. doi: 10.1016/j.tcb.2014.05.002. Epub 2014 Jun 18. Trends Cell Biol. 2014. PMID: 24953741 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM047337/GM/NIGMS NIH HHS/United States
- SRR022316A/PHS HHS/United States
- R21NS071166/NS/NINDS NIH HHS/United States
- 5-T32-DK07705/DK/NIDDK NIH HHS/United States
- GM47337/GM/NIGMS NIH HHS/United States
- 5P20RR017716-07/RR/NCRR NIH HHS/United States
- P20 RR017716/RR/NCRR NIH HHS/United States
- R21 NS071166/NS/NINDS NIH HHS/United States
- T32 DK007705/DK/NIDDK NIH HHS/United States
- S10 RR022316/RR/NCRR NIH HHS/United States
- R01 GM085306/GM/NIGMS NIH HHS/United States
- R01GM085306/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

