Comprehensive characterization of human genome variation by high coverage whole-genome sequencing of forty four Caucasians
- PMID: 23577066
- PMCID: PMC3618277
- DOI: 10.1371/journal.pone.0059494
Comprehensive characterization of human genome variation by high coverage whole-genome sequencing of forty four Caucasians
Abstract
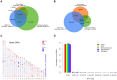
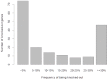
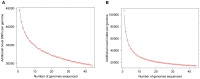
Whole genome sequencing studies are essential to obtain a comprehensive understanding of the vast pattern of human genomic variations. Here we report the results of a high-coverage whole genome sequencing study for 44 unrelated healthy Caucasian adults, each sequenced to over 50-fold coverage (averaging 65.8×). We identified approximately 11 million single nucleotide polymorphisms (SNPs), 2.8 million short insertions and deletions, and over 500,000 block substitutions. We showed that, although previous studies, including the 1000 Genomes Project Phase 1 study, have catalogued the vast majority of common SNPs, many of the low-frequency and rare variants remain undiscovered. For instance, approximately 1.4 million SNPs and 1.3 million short indels that we found were novel to both the dbSNP and the 1000 Genomes Project Phase 1 data sets, and the majority of which (∼96%) have a minor allele frequency less than 5%. On average, each individual genome carried ∼3.3 million SNPs and ∼492,000 indels/block substitutions, including approximately 179 variants that were predicted to cause loss of function of the gene products. Moreover, each individual genome carried an average of 44 such loss-of-function variants in a homozygous state, which would completely "knock out" the corresponding genes. Across all the 44 genomes, a total of 182 genes were "knocked-out" in at least one individual genome, among which 46 genes were "knocked out" in over 30% of our samples, suggesting that a number of genes are commonly "knocked-out" in general populations. Gene ontology analysis suggested that these commonly "knocked-out" genes are enriched in biological process related to antigen processing and immune response. Our results contribute towards a comprehensive characterization of human genomic variation, especially for less-common and rare variants, and provide an invaluable resource for future genetic studies of human variation and diseases.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

