Genome-level homology and phylogeny of Vibrionaceae (Gammaproteobacteria: Vibrionales) with three new complete genome sequences
- PMID: 23578061
- PMCID: PMC3663674
- DOI: 10.1186/1471-2180-13-80
Genome-level homology and phylogeny of Vibrionaceae (Gammaproteobacteria: Vibrionales) with three new complete genome sequences
Abstract
Background: Phylogenetic hypotheses based on complete genome data are presented for the Gammaproteobacteria family Vibrionaceae. Two taxon samplings are presented: one including all those taxa for which the genome sequences are complete in terms of arrangement (chromosomal location of fragments; 19 taxa) and one for which the genome sequences contain multiple contigs (44 taxa). Analyses are presented under the Maximum Parsimony and Maximum Likelihood optimality criteria for total evidence datasets, the two chromosomes separately, and individual analyses of locally collinear blocks. Three of the genomes included in the 44 taxon dataset, those of Vibrio gazogenes, Salinivibrio costicola, and Aliivibrio logei have been newly sequenced and their genome sequences are documented here.
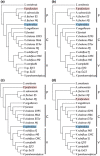
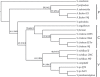
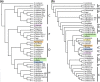
Results: Phylogenetic results for the 19-taxon datasets show similar levels of collinear subset of dataset incongruence as a previous study of 22 taxa from the sister family Shewanellaceae, while also echoing the strong phylogenetic performance of random subsets of data also shown in this study. Phylogenetic results for both the 19-taxon and 44-taxon datasets corroborate previous hypotheses about the placement of Photobacterium and Aliivibrio within Vibrionaceae and also highlight problems with how Photobacterium is delimited and indicate that it likely should be dissolved into Vibrio to produce a phylogenetic taxonomy. The 19-taxon and 44-taxon trees based on the large chromosome are congruent for the majority of taxa that are present in both datasets. Analyses of the 44-taxon sampling based on the second, small chromosome are quite different from those based on the large chromosome, which is not surprising given the dramatically divergent nature of the small chromosome and the difficulty in postulating primary homologies.
Conclusions: The phylogenetic analyses presented here represent the most comprehensive genome-level phylogenetic analyses in terms of taxa and data. Based on the availability of genome data for many bacterial species on GenBank, many other bacterial groups would also be amenable to similar genome-scale phylogenetic analyses even when present in multiple contigs. The result that collinear subsets of data are incongruent with the concatenated dataset and with each other while random data subsets show very little incongruence echoes the result of previous work on Shewanellaceae. The 44-taxon phylogenetic analysis presented here thus represents the future of phylogenomic analyses in scope and complexity.
Figures



Similar articles
-
Genome-level homology and phylogeny of Shewanella (Gammaproteobacteria: lteromonadales: Shewanellaceae).BMC Genomics. 2011 May 12;12:237. doi: 10.1186/1471-2164-12-237. BMC Genomics. 2011. PMID: 21569439 Free PMC article.
-
Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids.BMC Evol Biol. 2006 Apr 9;6:32. doi: 10.1186/1471-2148-6-32. BMC Evol Biol. 2006. PMID: 16603088 Free PMC article.
-
Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov.Int J Syst Evol Microbiol. 2007 Dec;57(Pt 12):2823-2829. doi: 10.1099/ijs.0.65081-0. Int J Syst Evol Microbiol. 2007. PMID: 18048732
-
Post-Genomic Analysis of Members of the Family Vibrionaceae.Microbiol Spectr. 2015 Oct;3(5):10.1128/microbiolspec.VE-0009-2014. doi: 10.1128/microbiolspec.VE-0009-2014. Microbiol Spectr. 2015. PMID: 26542048 Free PMC article. Review.
-
Vibrionaceae, a versatile bacterial family with evolutionarily conserved variability.Res Microbiol. 2007 Jul-Aug;158(6):479-86. doi: 10.1016/j.resmic.2007.04.007. Epub 2007 May 8. Res Microbiol. 2007. PMID: 17590316 Review.
Cited by
-
Independent origins and evolution of the secondary replicons of the class Gammaproteobacteria.Microb Genom. 2023 May;9(5):mgen001025. doi: 10.1099/mgen.0.001025. Microb Genom. 2023. PMID: 37185344 Free PMC article.
-
Vibrio chromosome-specific families.Front Microbiol. 2014 Mar 18;5:73. doi: 10.3389/fmicb.2014.00073. eCollection 2014. Front Microbiol. 2014. PMID: 24672511 Free PMC article.
-
The Pelagic Bacterium Paraphotobacterium marinum Has the Smallest Complete Genome Within the Family Vibrionaceae.Front Microbiol. 2017 Oct 11;8:1994. doi: 10.3389/fmicb.2017.01994. eCollection 2017. Front Microbiol. 2017. PMID: 29085348 Free PMC article.
-
Characterization of the Vibrio fischeri Fatty Acid Chemoreceptors, VfcB and VfcB2.Appl Environ Microbiol. 2015 Nov 13;82(2):696-704. doi: 10.1128/AEM.02856-15. Print 2016 Jan 15. Appl Environ Microbiol. 2015. PMID: 26567312 Free PMC article.
-
Parasalinivibrio latis gen. nov., sp. nov., isolated from the distal gut of healthy farmed Asian Seabass (Lates calcarifer).Antonie Van Leeuwenhoek. 2024 Nov 9;118(1):25. doi: 10.1007/s10482-024-02036-x. Antonie Van Leeuwenhoek. 2024. PMID: 39520647
References
-
- Naef A. Teuthologische notizen. 2. Gattungen Sepioliden. Zool Anz. 1912;39:244–248.
-
- Boisvert H, Chatelain R, Bassot JM. Etude d’un Photobacterium isole de l’organe lumineux des poissons Leiognathidae. Ann Inst Pasteur Paris. 1967;112:520–524. - PubMed
-
- Nesis KN. Cephalopods of the world. Neptune City: TFH Publications; 1982.
-
- Farmer JJI. International committee on systematic bacteriology. subcommittee on the taxonomy of Vibrionaceae. Minutes of the meetings. Int J Syst Bacteriol. 1986;39:210–12.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

