Self-assembling cages from coiled-coil peptide modules
- PMID: 23579496
- PMCID: PMC6485442
- DOI: 10.1126/science.1233936
Self-assembling cages from coiled-coil peptide modules
Abstract
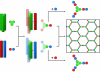
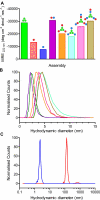
An ability to mimic the boundaries of biological compartments would improve our understanding of self-assembly and provide routes to new materials for the delivery of drugs and biologicals and the development of protocells. We show that short designed peptides can be combined to form unilamellar spheres approximately 100 nanometers in diameter. The design comprises two, noncovalent, heterodimeric and homotrimeric coiled-coil bundles. These are joined back to back to render two complementary hubs, which when mixed form hexagonal networks that close to form cages. This design strategy offers control over chemistry, self-assembly, reversibility, and size of such particles.
Figures


Comment in
-
Materials science. Obey the peptide assembly rules.Science. 2013 May 3;340(6132):561-2. doi: 10.1126/science.1237708. Science. 2013. PMID: 23641105 No abstract available.
-
Cages from coils.Nat Biotechnol. 2013 Sep;31(9):809-10. doi: 10.1038/nbt.2670. Nat Biotechnol. 2013. PMID: 24022157 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

