P4 ATPases: flippases in health and disease
- PMID: 23579954
- PMCID: PMC3645723
- DOI: 10.3390/ijms14047897
P4 ATPases: flippases in health and disease
Abstract
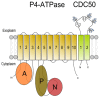
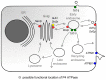
P4 ATPases catalyze the translocation of phospholipids from the exoplasmic to the cytosolic leaflet of biological membranes, a process termed "lipid flipping". Accumulating evidence obtained in lower eukaryotes points to an important role for P4 ATPases in vesicular protein trafficking. The human genome encodes fourteen P4 ATPases (fifteen in mouse) of which the cellular and physiological functions are slowly emerging. Thus far, deficiencies of at least two P4 ATPases, ATP8B1 and ATP8A2, are the cause of severe human disease. However, various mouse models and in vitro studies are contributing to our understanding of the cellular and physiological functions of P4-ATPases. This review summarizes current knowledge on the basic function of these phospholipid translocating proteins, their proposed action in intracellular vesicle transport and their physiological role.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

