Long-lasting alterations to DNA methylation and ncRNAs could underlie the effects of fetal alcohol exposure in mice
- PMID: 23580197
- PMCID: PMC3701217
- DOI: 10.1242/dmm.010975
Long-lasting alterations to DNA methylation and ncRNAs could underlie the effects of fetal alcohol exposure in mice
Abstract
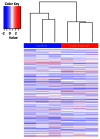
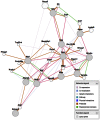
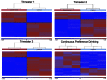
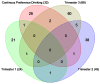
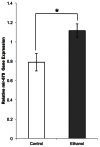
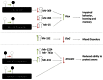
Fetal alcohol spectrum disorders (FASDs) are characterized by life-long changes in gene expression, neurodevelopment and behavior. What mechanisms initiate and maintain these changes are not known, but current research suggests a role for alcohol-induced epigenetic changes. In this study we assessed alterations to adult mouse brain tissue by assaying DNA cytosine methylation and small noncoding RNA (ncRNA) expression, specifically the microRNA (miRNA) and small nucleolar RNA (snoRNA) subtypes. We found long-lasting alterations in DNA methylation as a result of fetal alcohol exposure, specifically in the imprinted regions of the genome harboring ncRNAs and sequences interacting with regulatory proteins. A large number of major nodes from the identified networks, such as Pten signaling, contained transcriptional repressor CTCF-binding sites in their promoters, illustrating the functional consequences of alcohol-induced changes to DNA methylation. Next, we assessed ncRNA expression using two independent array platforms and quantitative PCR. The results identified 34 genes that are targeted by the deregulated miRNAs. Of these, four (Pten, Nmnat1, Slitrk2 and Otx2) were viewed as being crucial in the context of FASDs given their roles in the brain. Furthermore, ≈ 20% of the altered ncRNAs mapped to three imprinted regions (Snrpn-Ube3a, Dlk1-Dio3 and Sfmbt2) that showed differential methylation and have been previously implicated in neurodevelopmental disorders. The findings of this study help to expand on the mechanisms behind the long-lasting changes in the brain transcriptome of FASD individuals. The observed changes could contribute to the initiation and maintenance of the long-lasting effect of alcohol.
Figures

References
-
- Acampora D., Mazan S., Lallemand Y., Avantaggiato V., Maury M., Simeone A., Brûlet P. (1995). Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development 121, 3279–3290 - PubMed
-
- Allan A. M., Chynoweth J., Tyler L. A., Caldwell K. K. (2003). A mouse model of prenatal ethanol exposure using a voluntary drinking paradigm. Alcohol. Clin. Exp. Res. 27, 2009–2016 - PubMed
-
- Archibald S. L., Fennema-Notestine C., Gamst A., Riley E. P., Mattson S. N., Jernigan T. L. (2001). Brain dysmorphology in individuals with severe prenatal alcohol exposure. Dev. Med. Child Neurol. 43, 148–154 - PubMed
-
- Aruga J., Mikoshiba K. (2003). Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol. Cell. Neurosci. 24, 117–129 - PubMed
-
- Autti-Rämö I., Autti T., Korkman M., Kettunen S., Salonen O., Valanne L. (2002). MRI findings in children with school problems who had been exposed prenatally to alcohol. Dev. Med. Child Neurol. 44, 98–106 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

