PACo: a novel procrustes application to cophylogenetic analysis
- PMID: 23580325
- PMCID: PMC3620278
- DOI: 10.1371/journal.pone.0061048
PACo: a novel procrustes application to cophylogenetic analysis
Abstract
We present Procrustean Approach to Cophylogeny (PACo), a novel statistical tool to test for congruence between phylogenetic trees, or between phylogenetic distance matrices of associated taxa. Unlike previous tests, PACo evaluates the dependence of one phylogeny upon the other. This makes it especially appropriate to test the classical coevolutionary model that assumes that parasites that spend part of their life in or on their hosts track the phylogeny of their hosts. The new method does not require fully resolved phylogenies and allows for multiple host-parasite associations. PACo produces a Procrustes superimposition plot enabling a graphical assessment of the fit of the parasite phylogeny onto the host phylogeny and a goodness-of-fit statistic, whose significance is established by randomization of the host-parasite association data. The contribution of each individual host-parasite association to the global fit is measured by means of jackknife estimation of their respective squared residuals and confidence intervals associated to each host-parasite link. We carried out different simulations to evaluate the performance of PACo in terms of Type I and Type II errors with respect to two similar published tests. In most instances, PACo performed at least as well as the other tests and showed higher overall statistical power. In addition, the jackknife estimation of squared residuals enabled more elaborate validations about the nature of individual links than the ParaFitLink1 test of the program ParaFit. In order to demonstrate how it can be used in real biological situations, we applied PACo to two published studies using a script written in the public-domain statistical software R.
Conflict of interest statement
Figures
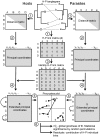
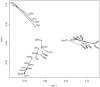
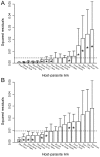
 ), whose significance can be established by a randomization procedure, and individual link residuals that can be further analysed to establish the contribution of each H-P link to the global fit.
), whose significance can be established by a randomization procedure, and individual link residuals that can be further analysed to establish the contribution of each H-P link to the global fit.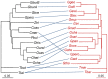

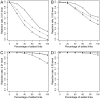
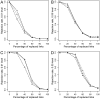


References
-
- Hafner MS, Demastes JW, Spradling TA, Reed DL (2003) Cophylogeny between pocket gophers and chewing lice. In: Page RDM, editor. Tangled Trees: Phylogeny, Cospeciation and Coevolution. Chicago: University of Chicago Press. 195–220.
-
- Jousselin E, Van Noort S, Berry V, Rasplus JY, Rønsted N, et al. (2008) One fig to bind them all: Host conservatism in a fig wasp community unraveled by cospeciation analyses among pollinating and nonpollinating fig wasps. Evolution 62: 1777–1797. - PubMed
-
- Baum BR, Johnson DA (2010) The application of cophylogenic tools to gene sets. Taxon 59: 1843–1852.
-
- Stevens J (2004) Computational aspects of host-parasite phylogenies. Briefings in bioinformatics 5: 339–349. - PubMed
-
- Ronquist F (1997) Phylogenetic approaches in coevolution and biogeography. Zoologica Scripta 26: 313–322.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

