Function relaxation followed by diversifying selection after whole-genome duplication in flowering plants
- PMID: 23580595
- PMCID: PMC3668069
- DOI: 10.1104/pp.112.213447
Function relaxation followed by diversifying selection after whole-genome duplication in flowering plants
Abstract
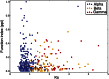
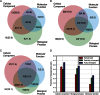
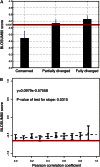
Episodes of whole-genome duplication (WGD) followed by gene loss dominate the evolutionary history of flowering plants. Despite the importance of understanding gene evolution following WGD, little is known about the evolutionary dynamics of this process. In this study, we analyzed duplicated genes from three WGD events in the Arabidopsis (Arabidopsis thaliana) lineage using multiple data types. Most duplicated genes that have survived from the most recent WGD (α) are under purifying selection in modern Arabidopsis populations. Using the number of identified protein-protein interactions as a proxy for functional divergence, approximately 92.7% of α-duplicated genes were diverged in function from one another in modern Arabidopsis populations, indicating that their preservation is no longer explicable by dosage balance. Dosage-balanced retention declines with antiquity of duplication: 24.1% of α-duplicated gene pairs in Arabidopsis remain in dosage balance with interacting partners, versus 12.9% and 9.4% for the earlier β-duplication and γ-triplication. GO-slim (a cut-down version of gene ontologies) terms reinforce evidence from protein-protein interactions, showing that the putatively diverged gene pairs are adapted to different cellular components. We identified a group of α-duplicated genes that show higher than average single-nucleotide polymorphism density, indicating that a period of positive selection, potentially driving functional divergence, may have preceded the current phase of purifying selection. We propose three possible paths for the evolution of duplicated genes following WGD.
Figures



Similar articles
-
Genome evolutionary dynamics followed by diversifying selection explains the complexity of the Sesamum indicum genome.BMC Genomics. 2017 Mar 24;18(1):257. doi: 10.1186/s12864-017-3599-4. BMC Genomics. 2017. PMID: 28340563 Free PMC article.
-
Extensive divergence in alternative splicing patterns after gene and genome duplication during the evolutionary history of Arabidopsis.Mol Biol Evol. 2010 Jul;27(7):1686-97. doi: 10.1093/molbev/msq054. Epub 2010 Feb 25. Mol Biol Evol. 2010. PMID: 20185454
-
Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants.Plant Physiol. 2013 Apr;161(4):1844-61. doi: 10.1104/pp.112.200147. Epub 2013 Feb 8. Plant Physiol. 2013. PMID: 23396833 Free PMC article.
-
Divergent evolutionary fates of major photosynthetic gene networks following gene and whole genome duplications.Plant Signal Behav. 2011 Apr;6(4):594-7. doi: 10.4161/psb.6.4.15370. Epub 2011 Apr 1. Plant Signal Behav. 2011. PMID: 21494088 Free PMC article. Review.
-
Insights into the structural and functional evolution of plant genomes afforded by the nucleotide sequences of chromosomes 2 and 4 of Arabidopsis thaliana.Yeast. 2000 Apr;17(1):1-5. doi: 10.1002/(SICI)1097-0061(200004)17:1<1::AID-YEA3>3.0.CO;2-V. Yeast. 2000. PMID: 10797596 Free PMC article. Review.
Cited by
-
Evolution of Gene Duplication in Plants.Plant Physiol. 2016 Aug;171(4):2294-316. doi: 10.1104/pp.16.00523. Epub 2016 Jun 10. Plant Physiol. 2016. PMID: 27288366 Free PMC article. Review.
-
Independent and Parallel Evolution of New Genes by Gene Duplication in Two Origins of C4 Photosynthesis Provides New Insight into the Mechanism of Phloem Loading in C4 Species.Mol Biol Evol. 2016 Jul;33(7):1796-806. doi: 10.1093/molbev/msw057. Epub 2016 Mar 24. Mol Biol Evol. 2016. PMID: 27016024 Free PMC article.
-
Resurrected Protein Interaction Networks Reveal the Innovation Potential of Ancient Whole-Genome Duplication.Plant Cell. 2018 Nov;30(11):2741-2760. doi: 10.1105/tpc.18.00409. Epub 2018 Oct 17. Plant Cell. 2018. PMID: 30333148 Free PMC article.
-
Chromosome-level genome assembly assisting for dissecting mechanism of anthocyanin regulation in kiwifruit (Actinidia arguta).Mol Hortic. 2025 Apr 1;5(1):18. doi: 10.1186/s43897-024-00139-7. Mol Hortic. 2025. PMID: 40165341 Free PMC article.
-
Gene duplication and genetic innovation in cereal genomes.Genome Res. 2019 Feb;29(2):261-269. doi: 10.1101/gr.237511.118. Epub 2019 Jan 16. Genome Res. 2019. PMID: 30651279 Free PMC article.
References
-
- Birchler JA, Riddle NC, Auger DL, Veitia RA. (2005) Dosage balance in gene regulation: biological implications. Trends Genet 21: 219–226 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

