Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3
- PMID: 23582324
- PMCID: PMC3661979
- DOI: 10.1016/j.cell.2013.03.012
Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3
Abstract
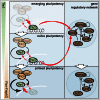
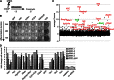
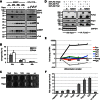
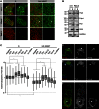
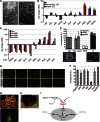
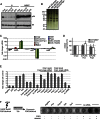
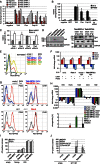
Factors that sustain self-renewal of mouse embryonic stem cells (ESCs) are well described. In contrast, the machinery regulating exit from pluripotency is ill defined. In a large-scale small interfering RNA (siRNA) screen, we found that knockdown of the tumor suppressors Folliculin (Flcn) and Tsc2 prevent ESC commitment. Tsc2 lies upstream of mammalian target of rapamycin (mTOR), whereas Flcn acts downstream and in parallel. Flcn with its interaction partners Fnip1 and Fnip2 drives differentiation by restricting nuclear localization and activity of the bHLH transcription factor Tfe3. Conversely, enforced nuclear Tfe3 enables ESCs to withstand differentiation conditions. Genome-wide location and functional analyses showed that Tfe3 directly integrates into the pluripotency circuitry through transcriptional regulation of Esrrb. These findings identify a cell-intrinsic rheostat for destabilizing ground-state pluripotency to allow lineage commitment. Congruently, stage-specific subcellular relocalization of Tfe3 suggests that Flcn-Fnip1/2 contributes to developmental progression of the pluripotent epiblast in vivo.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures








Comment in
-
Guards at the gate to embryonic stem cell differentiation.Cell. 2013 Apr 11;153(2):281-3. doi: 10.1016/j.cell.2013.03.037. Cell. 2013. PMID: 23582317
-
Stem cells: a gated exit from pluripotency.Nat Rev Mol Cell Biol. 2013 Jun;14(6):324. doi: 10.1038/nrm3585. Epub 2013 May 9. Nat Rev Mol Cell Biol. 2013. PMID: 23657495 No abstract available.
References
-
- Armah H.B., Parwani A.V. Xp11.2 translocation renal cell carcinoma. Arch. Pathol. Lab. Med. 2010;134:124–129. - PubMed
-
- Baba M., Hong S.B., Sharma N., Warren M.B., Nickerson M.L., Iwamatsu A., Esposito D., Gillette W.K., Hopkins R.F., 3rd, Hartley J.L. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc. Natl. Acad. Sci. USA. 2006;103:15552–15557. - PMC - PubMed
-
- Brannon M., Brown J.D., Bates R., Kimelman D., Moon R.T. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development. 1999;126:3159–3170. - PubMed
-
- Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A., Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. - PubMed
Supplemental References
-
- Baba M., Hong S.B., Sharma N., Warren M.B., Nickerson M.L., Iwamatsu A., Esposito D., Gillette W.K., Hopkins R.F., 3rd, Hartley J.L. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc. Natl. Acad. Sci. USA. 2006;103:15552–15557. - PMC - PubMed
-
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. - PubMed
-
- Hasumi Y., Baba M., Ajima R., Hasumi H., Valera V.A., Klein M.E., Haines D.C., Merino M.J., Hong S.B., Yamaguchi T.P. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc. Natl. Acad. Sci. USA. 2009;106:18722–18727. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

