Structure and T cell inhibition properties of B7 family member, B7-H3
- PMID: 23583036
- PMCID: PMC3998375
- DOI: 10.1016/j.str.2013.03.003
Structure and T cell inhibition properties of B7 family member, B7-H3
Abstract
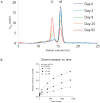
T cell activity is controlled by a combination of antigen-dependent signaling through the T cell receptor and a set of auxiliary signals delivered through antigen-independent interactions, including the recognition of the B7 family of ligands. B7-H3 is a recently identified B7 family member that is strongly overexpressed in a range of cancers and correlates with poor prognosis. We report the crystal structure of murine B7-H3 at a 3 Å resolution, which provides a model for the organization of the IgV and IgC domains within the ectodomain. We demonstrate that B7-H3 inhibits T cell proliferation and show that the FG loop of the IgV domain plays a critical role in this function. B7-H3 crystallized as an unusual dimer arising from the exchange of the G strands in the IgV domains of partner molecules. This arrangement, in combination with previous reports, highlights the dynamic nature and plasticity of the immunoglobulin fold.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


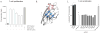

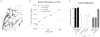

References
-
- Bennett MJ, Sawaya MR, Eisenberg D. Deposition diseases and 3D domain swapping. Structure. 2006;14:811–824. - PubMed
-
- Chapoval AI, Ni J, Lau JS, Wilcox RA, Flies DB, Liu D, Dong H, Sica GL, Zhu G, Tamada K, et al. B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol. 2001;2:269–274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30CA013330/CA/NCI NIH HHS/United States
- CA009173/CA/NCI NIH HHS/United States
- T32 CA009173/CA/NCI NIH HHS/United States
- GM094665/GM/NIGMS NIH HHS/United States
- GM094662/GM/NIGMS NIH HHS/United States
- R56 AI007289/AI/NIAID NIH HHS/United States
- P30 CA013330/CA/NCI NIH HHS/United States
- U54 GM094662/GM/NIGMS NIH HHS/United States
- DP2 DK083076/DK/NIDDK NIH HHS/United States
- AI007289/AI/NIAID NIH HHS/United States
- U01 GM094665/GM/NIGMS NIH HHS/United States
- DP2DK083076/DK/NIDDK NIH HHS/United States
- R01 AI007289/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

