Modeling autism by SHANK gene mutations in mice
- PMID: 23583105
- PMCID: PMC3659167
- DOI: 10.1016/j.neuron.2013.03.016
Modeling autism by SHANK gene mutations in mice
Abstract
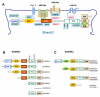
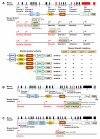
Shank family proteins (Shank1, Shank2, and Shank3) are synaptic scaffolding proteins that organize an extensive protein complex at the postsynaptic density (PSD) of excitatory glutamatergic synapses. Recent human genetic studies indicate that SHANK family genes (SHANK1, SHANK2, and SHANK3) are causative genes for idiopathic autism spectrum disorders (ASD). Neurobiological studies of Shank mutations in mice support a general hypothesis of synaptic dysfunction in the pathophysiology of ASD. However, the molecular diversity of SHANK family gene products, as well as the heterogeneity in human and mouse phenotypes, pose challenges to modeling human SHANK mutations. Here, we review the molecular genetics of SHANK mutations in human ASD and discuss recent findings where such mutations have been modeled in mice. Conserved features of synaptic dysfunction and corresponding behaviors in Shank mouse mutants may help dissect the pathophysiology of ASD, but also highlight divergent phenotypes that arise from different mutations in the same gene.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Translational neurobiology in Shank mutant mice--model systems for neuropsychiatric disorders.Ann Anat. 2015 Jul;200:115-7. doi: 10.1016/j.aanat.2015.03.006. Epub 2015 Apr 13. Ann Anat. 2015. PMID: 25917711 Review.
-
Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: a gradient of severity in cognitive impairments.PLoS Genet. 2014 Sep 4;10(9):e1004580. doi: 10.1371/journal.pgen.1004580. eCollection 2014 Sep. PLoS Genet. 2014. PMID: 25188300 Free PMC article.
-
SHANK1 and autism spectrum disorders.Sci China Life Sci. 2015 Oct;58(10):985-90. doi: 10.1007/s11427-015-4892-6. Sci China Life Sci. 2015. PMID: 26335738 Review.
-
SHANK1 polymorphisms and SNP-SNP interactions among SHANK family: A possible cue for recognition to autism spectrum disorder in infant age.Autism Res. 2019 Mar;12(3):375-383. doi: 10.1002/aur.2065. Epub 2019 Jan 10. Autism Res. 2019. PMID: 30629339
-
A role for synaptic zinc in ProSAP/Shank PSD scaffold malformation in autism spectrum disorders.Dev Neurobiol. 2014 Feb;74(2):136-46. doi: 10.1002/dneu.22089. Epub 2013 Sep 11. Dev Neurobiol. 2014. PMID: 23650259 Free PMC article. Review.
Cited by
-
Zinc Stabilizes Shank3 at the Postsynaptic Density of Hippocampal Synapses.PLoS One. 2016 May 4;11(5):e0153979. doi: 10.1371/journal.pone.0153979. eCollection 2016. PLoS One. 2016. PMID: 27144302 Free PMC article.
-
Behavioral and Sensory Deficits Associated with Dysfunction of GABAergic System in a Novel shank2-Deficient Zebrafish Model.Int J Mol Sci. 2023 Jan 22;24(3):2208. doi: 10.3390/ijms24032208. Int J Mol Sci. 2023. PMID: 36768529 Free PMC article.
-
Altered spinogenesis in iPSC-derived cortical neurons from patients with autism carrying de novo SHANK3 mutations.Sci Rep. 2019 Jan 14;9(1):94. doi: 10.1038/s41598-018-36993-x. Sci Rep. 2019. PMID: 30643170 Free PMC article.
-
Stem Cells from Human Exfoliated Deciduous Teeth Ameliorate Autistic-Like Behaviors of SHANK3 Mutant Beagle Dogs.Stem Cells Transl Med. 2022 Jul 20;11(7):778-789. doi: 10.1093/stcltm/szac028. Stem Cells Transl Med. 2022. PMID: 35608372 Free PMC article.
-
Abnormal Whisker-Dependent Behaviors and Altered Cortico-Hippocampal Connectivity in Shank3b-/- Mice.Cereb Cortex. 2022 Jul 12;32(14):3042-3056. doi: 10.1093/cercor/bhab399. Cereb Cortex. 2022. PMID: 34791077 Free PMC article.
References
-
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. - PubMed
-
- Anderlid BM, Schoumans J, Anneren G, Tapia-Paez I, Dumanski J, Blennow E, Nordenskjold M. FISH-mapping of a 100-kb terminal 22q13 deletion. Hum Genet. 2002;110:439–443. - PubMed
-
- Armstrong DD. Neuropathology of Rett syndrome. J Child Neurol. 2005;20:747–753. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

