Broadly neutralizing antibodies against influenza viruses
- PMID: 23583287
- PMCID: PMC3987986
- DOI: 10.1016/j.antiviral.2013.03.021
Broadly neutralizing antibodies against influenza viruses
Abstract
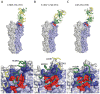
Despite available antivirals and vaccines, influenza continues to be a major cause of mortality worldwide. Vaccination generally induces an effective, but strain-specific antibody response. As the virus continually evolves, new vaccines have to be administered almost annually when a novel strain becomes dominant. Furthermore, the sporadic emerging resistance to neuraminidase inhibitors among circulating strains suggests an urgent need for new therapeutic agents. Recently, several cross-reactive antibodies have been described, which neutralize an unprecedented spectrum of influenza viruses. These broadly neutralizing antibodies generally target conserved functional regions on the major influenza surface glycoprotein hemagglutinin (HA). The characterization of their neutralization breadth and epitopes on HA could stimulate the development of new antibody-based antivirals and broader influenza vaccines. This article forms part of a symposium in Antiviral Research on "Treatment of influenza: targeting the virus or the host."
Copyright © 2013. Published by Elsevier B.V.
Conflict of interest statement
The authors have no financial or personal relationships that could be viewed as a potential conflict of interest.
Figures
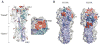


References
-
- Bommakanti G, Citron MP, Hepler RW, Callahan C, Heidecker GJ, Najar TA, Lu X, Joyce JG, Shiver JW, Casimiro DR, ter Meulen J, Liang X, Varadarajan R. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc Natl Acad Sci U S A. 2010;107:13701–13706. - PMC - PubMed
-
- Bright RA, Medina MJ, Xu X, Perez-Oronoz G, Wallis TR, Davis XM, Povinelli L, Cox NJ, Klimov AI. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–1181. - PubMed
-
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
-
- Carrat F, Flahault A. Influenza vaccine: the challenge of antigenic drift. Vaccine. 2007;25:6852–6862. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

