Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection
- PMID: 23583644
- PMCID: PMC3741679
- DOI: 10.1016/j.immuni.2013.02.020
Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection
Abstract
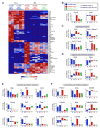
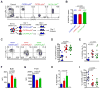
CD4(+) T follicular helper (Tfh) cells provide the required signals to B cells for germinal center reactions that are necessary for long-lived antibody responses. However, it remains unclear whether there are CD4(+) memory T cells committed to the Tfh cell lineage after antigen clearance. By using adoptive transfer of antigen-specific memory CD4(+) T cell subpopulations in the lymphocytic choriomeningitis virus infection model, we found that there are distinct memory CD4(+) T cell populations with commitment to either Tfh- or Th1-cell lineages. Our conclusions are based on gene expression profiles, epigenetic studies, and phenotypic and functional analyses. Our findings indicate that CD4(+) memory T cells "remember" their previous effector lineage after antigen clearance, being poised to reacquire their lineage-specific effector functions upon antigen reencounter. These findings have important implications for rational vaccine design, where improving the generation and engagement of memory Tfh cells could be used to enhance vaccine-induced protective immunity.
Copyright © 2013 Elsevier Inc. All rights reserved.
Conflict of interest statement
There are no financial conflicts of interest.
Figures




References
-
- Abdollahi A. LOT1 (ZAC/PLAGL1) and its family members: mechanisms and functions. Journal of Cellular Physiology. 2007;210:16–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

