Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold
- PMID: 23584677
- PMCID: PMC3661761
- DOI: 10.1038/nchembio.1231
Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold
Abstract
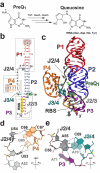
PreQ1 riboswitches regulate genes by binding the pyrrolopyrimidine intermediate preQ1 during the biosynthesis of the essential tRNA base queuosine. We report what is to our knowledge the first preQ1-II riboswitch structure at 2.3-Å resolution, which uses a previously uncharacterized fold to achieve effector recognition at the confluence of a three-way helical junction flanking a pseudoknotted ribosome-binding site. The results account for translational control mediated by the preQ1-II riboswitch class and expand the known repertoire of ligand-binding modes used by regulatory RNAs.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

