Identifying survival associated morphological features of triple negative breast cancer using multiple datasets
- PMID: 23585272
- PMCID: PMC3721170
- DOI: 10.1136/amiajnl-2012-001538
Identifying survival associated morphological features of triple negative breast cancer using multiple datasets
Abstract
Background and objective: Biomarkers for subtyping triple negative breast cancer (TNBC) are needed given the absence of responsive therapy and relatively poor prediction of survival. Morphology of cancer tissues is widely used in clinical practice for stratifying cancer patients, while genomic data are highly effective to classify cancer patients into subgroups. Thus integration of both morphological and genomic data is a promising approach in discovering new biomarkers for cancer outcome prediction. Here we propose a workflow for analyzing histopathological images and integrate them with genomic data for discovering biomarkers for TNBC.
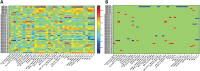
Materials and methods: We developed an image analysis workflow for extracting a large collection of morphological features and deployed the same on histological images from The Cancer Genome Atlas (TCGA) TNBC samples during the discovery phase (n=44). Strong correlations between salient morphological features and gene expression profiles from the same patients were identified. We then evaluated the same morphological features in predicting survival using a local TNBC cohort (n=143). We further tested the predictive power on patient prognosis of correlated gene clusters using two other public gene expression datasets.
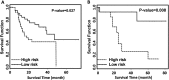
Results and conclusion: Using TCGA data, we identified 48 pairs of significantly correlated morphological features and gene clusters; four morphological features were able to separate the local cohort with significantly different survival outcomes. Gene clusters correlated with these four morphological features further proved to be effective in predicting patient survival using multiple public gene expression datasets. These results suggest the efficacy of our workflow and demonstrate that integrative analysis holds promise for discovering biomarkers of complex diseases.
Keywords: Biomarker Identification; Cancer Survival; Computational Biology; Image Analysis; The Cancer Genome Atlas; Triple Negative Breast Cancer.
Figures




References
-
- Irshad S, Ellis P, Tutt A. Molecular heterogeneity of triple-negative breast cancer and its clinical implications. Curr Opin Oncol 2011;23:566–77 - PubMed
-
- Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist 2011;16:61–70 - PubMed
-
- Nafe R, Franz K, Schlote W, et al. Morphology of tumor cell nuclei is significantly related with survival time of patients with glioblastomas. Clin Cancer Res 2005;11:2141–8 - PubMed
-
- Khan OA, Fitzgerald JJ, Field ML, et al. Histological determinants of survival in completely resected T1-2N1M0 nonsmall cell cancer of the lung. Ann Thorac Surg 2004;77:1173–8 - PubMed
-
- Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med 2011;3:108–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

