Zebrafish AID is capable of deaminating methylated deoxycytidines
- PMID: 23585279
- PMCID: PMC3664802
- DOI: 10.1093/nar/gkt212
Zebrafish AID is capable of deaminating methylated deoxycytidines
Abstract
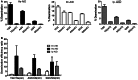
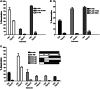
Activation-induced cytidine deaminase (AID) deaminates deoxycytidine (dC) to deoxyuracil (dU) at immunoglobulin loci in B lymphocytes to mediate secondary antibody diversification. Recently, AID has been proposed to also mediate epigenetic reprogramming by demethylating methylated cytidines (mC) possibly through deamination. AID overexpression in zebrafish embryos was shown to promote genome demethylation through G:T lesions, implicating a deamination-dependent mechanism. We and others have previously shown that mC is a poor substrate for human AID. Here, we examined the ability of bony fish AID to deaminate mC. We report that zebrafish AID was unique among all orthologs in that it efficiently deaminates mC. Analysis of domain-swapped and mutant AID revealed that mC specificity is independent of the overall high-catalytic efficiency of zebrafish AID. Structural modeling with or without bound DNA suggests that efficient deamination of mC by zebrafish AID is likely not due to a larger catalytic pocket allowing for better fit of mC, but rather because of subtle differences in the flexibility of its structure.
Figures

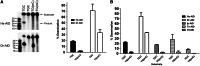


References
-
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. - PubMed
-
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. - PubMed
-
- Larijani M, Frieder D, Basit W, Martin A. The mutation spectrum of purified AID is similar to the mutability index in Ramos cells and in ung(−/−)msh2(−/−) mice. Immunogenetics. 2005;56:840–845. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

