Mass spectrometry-based proteomics in molecular diagnostics: discovery of cancer biomarkers using tissue culture
- PMID: 23586059
- PMCID: PMC3613068
- DOI: 10.1155/2013/783131
Mass spectrometry-based proteomics in molecular diagnostics: discovery of cancer biomarkers using tissue culture
Abstract
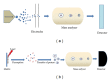
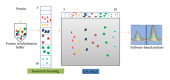
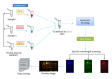
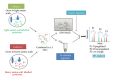
Accurate diagnosis and proper monitoring of cancer patients remain a key obstacle for successful cancer treatment and prevention. Therein comes the need for biomarker discovery, which is crucial to the current oncological and other clinical practices having the potential to impact the diagnosis and prognosis. In fact, most of the biomarkers have been discovered utilizing the proteomics-based approaches. Although high-throughput mass spectrometry-based proteomic approaches like SILAC, 2D-DIGE, and iTRAQ are filling up the pitfalls of the conventional techniques, still serum proteomics importunately poses hurdle in overcoming a wide range of protein concentrations, and also the availability of patient tissue samples is a limitation for the biomarker discovery. Thus, researchers have looked for alternatives, and profiling of candidate biomarkers through tissue culture of tumor cell lines comes up as a promising option. It is a rich source of tumor cell-derived proteins, thereby, representing a wide array of potential biomarkers. Interestingly, most of the clinical biomarkers in use today (CA 125, CA 15.3, CA 19.9, and PSA) were discovered through tissue culture-based system and tissue extracts. This paper tries to emphasize the tissue culture-based discovery of candidate biomarkers through various mass spectrometry-based proteomic approaches.
Figures

References
-
- Etzioni R, Urban N, Ramsey S, et al. The case for early detection. Nature Reviews Cancer. 2003;3(4):243–252. - PubMed
-
- Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nature Clinical Practice Oncology. 2008;5(10):588–599. - PubMed
-
- Hayes DF, Bast RC, Desch CE, et al. Tumor marker utility grading system: a framework to evaluate clinical utility of tumor markers. Journal of the National Cancer Institute. 1996;88(20):1456–1466. - PubMed
-
- Minamida S, Iwamura M, Kodera Y, et al. Profilin 1 overexpression in renal cell carcinoma. International Journal of Urology. 2011;18(1):63–71. - PubMed
-
- Xu X, Qiao M, Zhang Y, et al. Quantitative proteomics study of breast cancer cell lines isolated from a single patient: discovery of TIMM17A as a marker for breast cancer. Proteomics. 2010;10(7):1374–1390. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

