A HuD-ZBP1 ribonucleoprotein complex localizes GAP-43 mRNA into axons through its 3' untranslated region AU-rich regulatory element
- PMID: 23586486
- PMCID: PMC3766383
- DOI: 10.1111/jnc.12266
A HuD-ZBP1 ribonucleoprotein complex localizes GAP-43 mRNA into axons through its 3' untranslated region AU-rich regulatory element
Abstract
Localized translation of axonal mRNAs contributes to developmental and regenerative axon growth. Although untranslated regions (UTRs) of many different axonal mRNAs appear to drive their localization, there has been no consensus RNA structure responsible for this localization. We recently showed that limited expression of ZBP1 protein restricts axonal localization of both β-actin and GAP-43 mRNAs. β-actin 3'UTR has a defined element for interaction with ZBP1, but GAP-43 mRNA shows no homology to this RNA sequence. Here, we show that an AU-rich regulatory element (ARE) in GAP-43's 3'UTR is necessary and sufficient for its axonal localization. Axonal GAP-43 mRNA levels increase after in vivo injury, and GAP-43 mRNA shows an increased half-life in regenerating axons. GAP-43 mRNA interacts with both HuD and ZBP1, and HuD and ZBP1 co-immunoprecipitate in an RNA-dependent fashion. Reporter mRNA with the GAP-43 ARE competes with endogenous β-actin mRNA for axonal localization and decreases axon length and branching similar to the β-actin 3'UTR competing with endogenous GAP-43 mRNA. Conversely, over-expressing GAP-43 coding sequence with its 3'UTR ARE increases axonal elongation and this effect is lost when just the ARE is deleted from GAP-43's 3'UTR. We have recently found that over-expression of GAP-43 using an axonally targeted construct with the 3'UTRs of GAP-43 promoted elongating growth of axons, while restricting the mRNA to the cell body with the 3'UTR of γ-actin had minimal effect on axon length. In this study, we show that the ARE in GAP-43's 3'UTR is responsible for localization of GAP-43 mRNA into axons and is sufficient for GAP-43 protein's role in elongating axonal growth.
Keywords: GAP-43; HuD; RNA-immunoprecipitation; ZBP1; axon regeneration; mRNA transport.
© 2013 International Society for Neurochemistry.
Figures



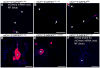
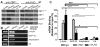
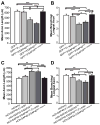
References
-
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. - PubMed
-
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–474. - PubMed
-
- Aronov S, Marx R, Ginzburg I. Identification of 3′UTR region implicated in tau mRNA stabilization in neuronal cells. J Mol Neurosci. 1999;12:131–145. - PubMed
-
- Atlas R, Behar L, Elliott E, Ginzburg I. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J Neurochem. 2004;89:613–626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

