Quantitative imaging biomarker ontology (QIBO) for knowledge representation of biomedical imaging biomarkers
- PMID: 23589184
- PMCID: PMC3705004
- DOI: 10.1007/s10278-013-9599-2
Quantitative imaging biomarker ontology (QIBO) for knowledge representation of biomedical imaging biomarkers
Erratum in
- J Digit Imaging. 2013 Aug;26(4):642. Ouellette, M [removed]; Danagoulian, J [removed]; Wernsing, G [removed]
Abstract
A widening array of novel imaging biomarkers is being developed using ever more powerful clinical and preclinical imaging modalities. These biomarkers have demonstrated effectiveness in quantifying biological processes as they occur in vivo and in the early prediction of therapeutic outcomes. However, quantitative imaging biomarker data and knowledge are not standardized, representing a critical barrier to accumulating medical knowledge based on quantitative imaging data. We use an ontology to represent, integrate, and harmonize heterogeneous knowledge across the domain of imaging biomarkers. This advances the goal of developing applications to (1) improve precision and recall of storage and retrieval of quantitative imaging-related data using standardized terminology; (2) streamline the discovery and development of novel imaging biomarkers by normalizing knowledge across heterogeneous resources; (3) effectively annotate imaging experiments thus aiding comprehension, re-use, and reproducibility; and (4) provide validation frameworks through rigorous specification as a basis for testable hypotheses and compliance tests. We have developed the Quantitative Imaging Biomarker Ontology (QIBO), which currently consists of 488 terms spanning the following upper classes: experimental subject, biological intervention, imaging agent, imaging instrument, image post-processing algorithm, biological target, indicated biology, and biomarker application. We have demonstrated that QIBO can be used to annotate imaging experiments with standardized terms in the ontology and to generate hypotheses for novel imaging biomarker-disease associations. Our results established the utility of QIBO in enabling integrated analysis of quantitative imaging data.
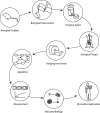
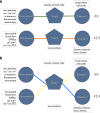
Figures





References
-
- Buckler AJ, Boellaard R. Standardization of quantitative imaging: the time is right, and 18F-FDG PET/CT is a good place to start. Journal of Nuclear Medicine: Official Publication, Society of Nuclear Medicine. 2011;52(2):171–172. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

