Clinical analysis and interpretation of cancer genome data
- PMID: 23589549
- PMCID: PMC4878102
- DOI: 10.1200/JCO.2013.48.7215
Clinical analysis and interpretation of cancer genome data
Abstract
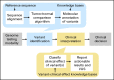
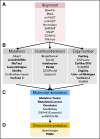
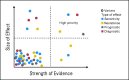
The scale of tumor genomic profiling is rapidly outpacing human cognitive capacity to make clinical decisions without the aid of tools. New frameworks are needed to help researchers and clinicians process the information emerging from the explosive growth in both the number of tumor genetic variants routinely tested and the respective knowledge to interpret their clinical significance. We review the current state, limitations, and future trends in methods to support the clinical analysis and interpretation of cancer genomes. This includes the processes of genome-scale variant identification, including tools for sequence alignment, tumor-germline comparison, and molecular annotation of variants. The process of clinical interpretation of tumor variants includes classification of the effect of the variant, reporting the results to clinicians, and enabling the clinician to make a clinical decision based on the genomic information integrated with other clinical features. We describe existing knowledge bases, databases, algorithms, and tools for identification and visualization of tumor variants and their actionable subsets. With the decreasing cost of tumor gene mutation testing and the increasing number of actionable therapeutics, we expect the methods for analysis and interpretation of cancer genomes to continue to evolve to meet the needs of patient-centered clinical decision making. The science of computational cancer medicine is still in its infancy; however, there is a clear need to continue the development of knowledge bases, best practices, tools, and validation experiments for successful clinical implementation in oncology.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

