S1-S3 counter charges in the voltage sensor module of a mammalian sodium channel regulate fast inactivation
- PMID: 23589580
- PMCID: PMC3639575
- DOI: 10.1085/jgp.201210935
S1-S3 counter charges in the voltage sensor module of a mammalian sodium channel regulate fast inactivation
Erratum in
-
Correction: S1-S3 counter charges in the voltage sensor module of a mammalian sodium channel regulate fast inactivation.J Gen Physiol. 2015 Dec;146(6):541-6. doi: 10.1085/jgp.20121093510232015c. Epub 2015 Nov 16. J Gen Physiol. 2015. PMID: 26573623 Free PMC article. No abstract available.
Abstract
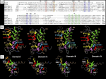
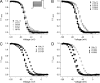
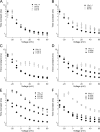
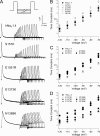
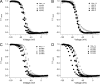
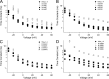
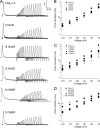
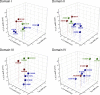
The movement of positively charged S4 segments through the electric field drives the voltage-dependent gating of ion channels. Studies of prokaryotic sodium channels provide a mechanistic view of activation facilitated by electrostatic interactions of negatively charged residues in S1 and S2 segments, with positive counterparts in the S4 segment. In mammalian sodium channels, S4 segments promote domain-specific functions that include activation and several forms of inactivation. We tested the idea that S1-S3 countercharges regulate eukaryotic sodium channel functions, including fast inactivation. Using structural data provided by bacterial channels, we constructed homology models of the S1-S4 voltage sensor module (VSM) for each domain of the mammalian skeletal muscle sodium channel hNaV1.4. These show that side chains of putative countercharges in hNaV1.4 are oriented toward the positive charge complement of S4. We used mutagenesis to define the roles of conserved residues in the extracellular negative charge cluster (ENC), hydrophobic charge region (HCR), and intracellular negative charge cluster (INC). Activation was inhibited with charge-reversing VSM mutations in domains I-III. Charge reversal of ENC residues in domains III (E1051R, D1069K) and IV (E1373K, N1389K) destabilized fast inactivation by decreasing its probability, slowing entry, and accelerating recovery. Several INC mutations increased inactivation from closed states and slowed recovery. Our results extend the functional characterization of VSM countercharges to fast inactivation, and support the premise that these residues play a critical role in domain-specific gating transitions for a mammalian sodium channel.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

