Claudins and the modulation of tight junction permeability
- PMID: 23589827
- PMCID: PMC3768107
- DOI: 10.1152/physrev.00019.2012
Claudins and the modulation of tight junction permeability
Abstract
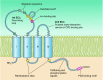
Claudins are tight junction membrane proteins that are expressed in epithelia and endothelia and form paracellular barriers and pores that determine tight junction permeability. This review summarizes our current knowledge of this large protein family and discusses recent advances in our understanding of their structure and physiological functions.
Figures


References
-
- Acharya P, Beckel J, Ruiz WG, Wang E, Rojas R, Birder L, Apodaca G. Distribution of the tight junction proteins ZO-1, occludin, claudin-4, -8, and -12 in bladder epithelium. Am J Physiol Renal Physiol 287: F305–F318, 2004 - PubMed
-
- Aijaz S, Balda MS, Matter K. Tight junctions: molecular architecture and function. Int Rev Cytol 248: 261–298, 2006 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

