Protein-DNA complexes are the primary sources of replication fork pausing in Escherichia coli
- PMID: 23589869
- PMCID: PMC3645559
- DOI: 10.1073/pnas.1303890110
Protein-DNA complexes are the primary sources of replication fork pausing in Escherichia coli
Abstract
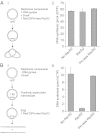
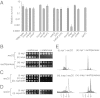
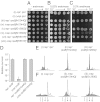
Replication fork pausing drives genome instability, because any loss of paused replisome activity creates a requirement for reloading of the replication machinery, a potentially mutagenic process. Despite this importance, the relative contributions to fork pausing of different replicative barriers remain unknown. We show here that Deinococcus radiodurans RecD2 helicase inactivates Escherichia coli replisomes that are paused but still functional in vitro, preventing continued fork movement upon barrier removal or bypass, but does not inactivate elongating forks. Using RecD2 to probe replisome pausing in vivo, we demonstrate that most pausing events do not lead to replisome inactivation, that transcription complexes are the primary sources of this pausing, and that an accessory replicative helicase is critical for minimizing the frequency and/or duration of replisome pauses. These findings reveal the hidden potential for replisome inactivation, and hence genome instability, inside cells. They also demonstrate that efficient chromosome duplication requires mechanisms that aid resumption of replication by paused replisomes, especially those halted by protein-DNA barriers such as transcription complexes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992;258(5086):1362–1365. - PubMed
-
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286(5446):1897–1905. - PubMed
-
- Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7(12):932–943. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0800970/MRC_/Medical Research Council/United Kingdom
- BB/I001859/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- GM34557/GM/NIGMS NIH HHS/United States
- BB/G005915/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R37 GM034557/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

