Genetic diversity of bile salt hydrolases among human intestinal bifidobacteria
- PMID: 23591474
- PMCID: PMC3722454
- DOI: 10.1007/s00284-013-0362-1
Genetic diversity of bile salt hydrolases among human intestinal bifidobacteria
Abstract
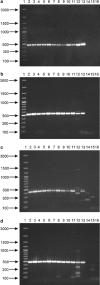
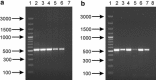
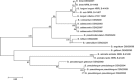
This study analyzes the application of degenerative primers for the screening of bile salt hydrolase-encoding genes (bsh) in various intestinal bifidobacteria. In the first stage, the design and evaluation of the universal PCR primers for amplifying the partial coding sequence of bile salt hydrolase in bifidobacteria were performed. The amplified bsh gene fragments were sequenced and the obtained sequences were compared to the bsh genes present in GenBank. The determined results showed the utility of the designed PCR primers for the amplification of partial gene encoding bile salt hydrolase in different intestinal bifidobacteria. Moreover, sequence analysis revealed that bile salt hydrolase-encoding genes may be used as valuable molecular markers for phylogenetic studies and identification of even closely related members of the genus Bifidobacterium.
Figures

References
-
- Alschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

