DNA self-assembly: from chirality to evolution
- PMID: 23591841
- PMCID: PMC3645741
- DOI: 10.3390/ijms14048252
DNA self-assembly: from chirality to evolution
Abstract
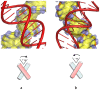
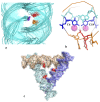
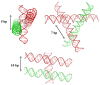
Transient or long-term DNA self-assembly participates in essential genetic functions. The present review focuses on tight DNA-DNA interactions that have recently been found to play important roles in both controlling DNA higher-order structures and their topology. Due to their chirality, double helices are tightly packed into stable right-handed crossovers. Simple packing rules that are imposed by DNA geometry and sequence dictate the overall architecture of higher order DNA structures. Close DNA-DNA interactions also provide the missing link between local interactions and DNA topology, thus explaining how type II DNA topoisomerases may sense locally the global topology. Finally this paper proposes that through its influence on DNA self-assembled structures, DNA chirality played a critical role during the early steps of evolution.
Figures


References
-
- Echols H. Nucleoprotein structures initiating DNA-replication, transcription, and site-specific recombination. J. Biol. Chem. 1990;265:14697–14700. - PubMed
-
- Grosschedl R. Higher-order nucleoprotein complexes in transcription—Analogies with site-specific recombination. Curr. Opin. Cell Biol. 1995;7:362–370. - PubMed
-
- Bloomfield V.A. DNA condensation. Curr. Opin. Struct. Biol. 1996;6:334–341. - PubMed
-
- Strey H.H., Podgornik R., Rau D.C., Parsegian V.A. DNA-DNA interactions. Curr. Opin. Struct. Biol. 1998;8:309–313. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

