Tertiary model of a plant cellulose synthase
- PMID: 23592721
- PMCID: PMC3645513
- DOI: 10.1073/pnas.1301027110
Tertiary model of a plant cellulose synthase
Abstract
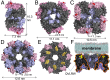
A 3D atomistic model of a plant cellulose synthase (CESA) has remained elusive despite over forty years of experimental effort. Here, we report a computationally predicted 3D structure of 506 amino acids of cotton CESA within the cytosolic region. Comparison of the predicted plant CESA structure with the solved structure of a bacterial cellulose-synthesizing protein validates the overall fold of the modeled glycosyltransferase (GT) domain. The coaligned plant and bacterial GT domains share a six-stranded β-sheet, five α-helices, and conserved motifs similar to those required for catalysis in other GT-2 glycosyltransferases. Extending beyond the cross-kingdom similarities related to cellulose polymerization, the predicted structure of cotton CESA reveals that plant-specific modules (plant-conserved region and class-specific region) fold into distinct subdomains on the periphery of the catalytic region. Computational results support the importance of the plant-conserved region and/or class-specific region in CESA oligomerization to form the multimeric cellulose-synthesis complexes that are characteristic of plants. Relatively high sequence conservation between plant CESAs allowed mapping of known mutations and two previously undescribed mutations that perturb cellulose synthesis in Arabidopsis thaliana to their analogous positions in the modeled structure. Most of these mutation sites are near the predicted catalytic region, and the confluence of other mutation sites supports the existence of previously undefined functional nodes within the catalytic core of CESA. Overall, the predicted tertiary structure provides a platform for the biochemical engineering of plant CESAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
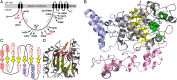


References
-
- Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. - PubMed
-
- Nishiyama Y. Structure and properties of the cellulose microfi bril. J Wood Sci. 2009;55:241–249.
-
- Roberts E, Roberts AW. A cellulose synthase (Cesa) gene from the red alga Porphyra yezoensis (Rhodophyta) J Phycol. 2009;45:203–212. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

