In vivo live imaging of RNA polymerase II transcription factories in primary cells
- PMID: 23592796
- PMCID: PMC3639417
- DOI: 10.1101/gad.216200.113
In vivo live imaging of RNA polymerase II transcription factories in primary cells
Erratum in
- Genes Dev. 2013 Jun 15;27(12):1434
- Genes Dev. 2013 May 15;27(10):1216
Abstract
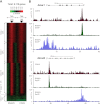
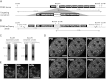
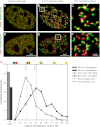
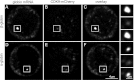
Transcription steps are marked by different modifications of the C-terminal domain of RNA polymerase II (RNAPII). Phosphorylation of Ser5 and Ser7 by cyclin-dependent kinase 7 (CDK7) as part of TFIIH marks initiation, whereas phosphorylation of Ser2 by CDK9 marks elongation. These processes are thought to take place in localized transcription foci in the nucleus, known as "transcription factories," but it has been argued that the observed clusters/foci are mere fixation or labeling artifacts. We show that transcription factories exist in living cells as distinct foci by live-imaging fluorescently labeled CDK9, a kinase known to associate with active RNAPII. These foci were observed in different cell types derived from CDK9-mCherry knock-in mice. We show that these foci are very stable while highly dynamic in exchanging CDK9. Chromatin immunoprecipitation (ChIP) coupled with deep sequencing (ChIP-seq) data show that the genome-wide binding sites of CDK9 and initiating RNAPII overlap on transcribed genes. Immunostaining shows that CDK9-mCherry foci colocalize with RNAPII-Ser5P, much less with RNAPII-Ser2P, and not with CDK12 (a kinase reported to be involved in the Ser2 phosphorylation) or with splicing factor SC35. In conclusion, transcription factories exist in living cells, and initiation and elongation of transcripts takes place in different nuclear compartments.
Figures


References
-
- Chandler D 2005. Interfaces and the driving force of hydrophobic assembly. Nature 437: 640–647 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
