Nutrient signaling in protein homeostasis: an increase in quantity at the expense of quality
- PMID: 23592839
- PMCID: PMC3992710
- DOI: 10.1126/scisignal.2003520
Nutrient signaling in protein homeostasis: an increase in quantity at the expense of quality
Abstract
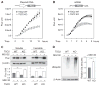
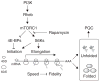
The discovery that rapamycin extends the life span of diverse organisms has triggered many studies aimed at identifying the underlying molecular mechanisms. Mammalian target of rapamycin complex 1 (mTORC1) regulates cell growth and may regulate organismal aging by controlling mRNA translation. However, how inhibiting mTORC1 and decreasing protein synthesis can extend life span remains an unresolved issue. We showed that constitutively active mTORC1 signaling increased general protein synthesis but unexpectedly reduced the quality of newly synthesized polypeptides. We demonstrated that constitutively active mTORC1 decreased translation fidelity by increasing the speed of ribosomal elongation. Conversely, rapamycin treatment restored the quality of newly synthesized polypeptides mainly by slowing the rate of ribosomal elongation. We also found distinct roles for mTORC1 downstream targets in maintaining protein homeostasis. Loss of S6 kinases, but not 4E-BP family proteins, which are both involved in regulation of translation, attenuated the effects of rapamycin on the quality of newly translated proteins. Our results reveal a mechanistic connection between mTORC1 and protein quality, highlighting the central role of nutrient signaling in growth and aging.
Conflict of interest statement
Figures




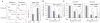

References
-
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. - PubMed
-
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. - PubMed
-
- Inoki K, Guan KL. Complexity of the TOR signaling network. Trends Cell Biol. 2006;16:206–212. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

