Site-specific phosphorylation of the DNA damage response mediator rad9 by cyclin-dependent kinases regulates activation of checkpoint kinase 1
- PMID: 23593009
- PMCID: PMC3616908
- DOI: 10.1371/journal.pgen.1003310
Site-specific phosphorylation of the DNA damage response mediator rad9 by cyclin-dependent kinases regulates activation of checkpoint kinase 1
Abstract
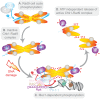
The mediators of the DNA damage response (DDR) are highly phosphorylated by kinases that control cell proliferation, but little is known about the role of this regulation. Here we show that cell cycle phosphorylation of the prototypical DDR mediator Saccharomyces cerevisiae Rad9 depends on cyclin-dependent kinase (CDK) complexes. We find that a specific G2/M form of Cdc28 can phosphorylate in vitro the N-terminal region of Rad9 on nine consensus CDK phosphorylation sites. We show that the integrity of CDK consensus sites and the activity of Cdc28 are required for both the activation of the Chk1 checkpoint kinase and its interaction with Rad9. We have identified T125 and T143 as important residues in Rad9 for this Rad9/Chk1 interaction. Phosphorylation of T143 is the most important feature promoting Rad9/Chk1 interaction, while the much more abundant phosphorylation of the neighbouring T125 residue impedes the Rad9/Chk1 interaction. We suggest a novel model for Chk1 activation where Cdc28 regulates the constitutive interaction of Rad9 and Chk1. The Rad9/Chk1 complex is then recruited at sites of DNA damage where activation of Chk1 requires additional DDR-specific protein kinases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745. - PubMed
-
- Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355. - PubMed
-
- Durocher D, Jackson SP (2001) DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr Opin Cell Biol 13: 225–231. - PubMed
-
- FitzGerald JE, Grenon M, Lowndes NF (2009) 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans 37: 897–904. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

